1Q77
 
 | | X-ray crystal structure of putative Universal Stress Protein from Aquifex aeolicus | | Descriptor: | Hypothetical protein AQ_178, SULFATE ION | | Authors: | Osipiuk, J, Zhou, M, Moy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-08-16 | | Release date: | 2003-11-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural homolog of Universal Stress Protein from Aquifex aeolicus
To be Published
|
|
3KRV
 
 | | The Structure Of Potential Metal-Dependent Hydrolase With Cyclase Activity | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Rakonjac, N, Rezacova, P, Borek, D, Collart, F, Joachimiak, A, Otwinowski, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-11-19 | | Release date: | 2010-01-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The Structure Of Potential Metal-Dependent Hydrolase With Cyclase Activity
To be Published
|
|
2AEE
 
 | | Crystal structure of Orotate phosphoribosyltransferase from Streptococcus pyogenes | | Descriptor: | CHLORIDE ION, GLYCEROL, Orotate phosphoribosyltransferase, ... | | Authors: | Chang, C, Li, H, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-07-22 | | Release date: | 2005-09-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of Orotate phosphoribosyltransferase from Streptococcus pyogenes
To be Published
|
|
2AE8
 
 | | Crystal Structure of Imidazoleglycerol-phosphate Dehydratase from Staphylococcus aureus subsp. aureus N315 | | Descriptor: | Imidazoleglycerol-phosphate dehydratase, MAGNESIUM ION | | Authors: | Kim, Y, Quartey, P, Holzle, D, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-07-21 | | Release date: | 2005-09-06 | | Last modified: | 2015-05-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structure of Imidazoleglycerol-phosphate Dehydratase from Staphylococcus aureus subsp. aureus N315
To be Published
|
|
2A67
 
 | | Crystal structure of Isochorismatase family protein | | Descriptor: | isochorismatase family protein | | Authors: | Chang, C, Hatzos, C, Collart, F, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-07-01 | | Release date: | 2005-08-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Isochorismatase family protein
To be Published
|
|
2AE6
 
 | | Crystal Structure of Acetyltransferase of GNAT family from Enterococcus faecalis V583 | | Descriptor: | ETHANOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kim, Y, Hatzos, C, Moy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-07-21 | | Release date: | 2005-09-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of Acetyltransferase of GNAT family from Enterococcus faecalis V583
To be Published
|
|
2AO9
 
 | |
1I60
 
 | | Structural genomics, IOLI protein | | Descriptor: | IOLI PROTEIN | | Authors: | Zhang, R, Dementieva, I, Collart, F, Quaite-Randall, E, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-03-01 | | Release date: | 2002-03-13 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Bacillus subtilis ioli shows endonuclase IV fold with altered Zn binding.
Proteins, 48, 2002
|
|
1I6N
 
 | | 1.8 A Crystal structure of IOLI protein with a binding zinc atom | | Descriptor: | IOLI PROTEIN, ZINC ION | | Authors: | Zhang, R.G, Dementiva, I, Collart, F, Quaite-Randall, E, Joachimiak, A, Alkire, R, Maltsev, N, Korolev, O, Dieckman, L, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-03-02 | | Release date: | 2002-03-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Bacillus subtilis ioli shows endonuclase IV fold with altered Zn binding.
Proteins, 48, 2002
|
|
1K6D
 
 | | CRYSTAL STRUCTURE OF ACETATE COA-TRANSFERASE ALPHA SUBUNIT | | Descriptor: | ACETATE COA-TRANSFERASE ALPHA SUBUNIT, MAGNESIUM ION | | Authors: | Korolev, S, Koroleva, O, Petterson, K, Collart, F, Dementieva, I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-10-15 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Autotracing of Escherichia coli acetate CoA-transferase alpha-subunit structure using 3.4 A MAD and 1.9 A native data.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1KYH
 
 | | Structure of Bacillus subtilis YxkO, a Member of the UPF0031 Family and a Putative Kinase | | Descriptor: | Hypothetical 29.9 kDa protein in SIGY-CYDD intergenic region | | Authors: | Zhang, R, Dementieva, I, Vinokour, E, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-02-04 | | Release date: | 2002-08-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Bacillus subtilis YXKO--a member of the UPF0031 family and a putative kinase.
J.Struct.Biol., 139, 2002
|
|
2OEQ
 
 | |
2OSU
 
 | | Probable glutaminase from Bacillus subtilis complexed with 6-diazo-5-oxo-L-norleucine | | Descriptor: | 6-DIAZENYL-5-OXO-L-NORLEUCINE, Glutaminase 1 | | Authors: | Kim, Y, Dementieva, I, Vinokour, E, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-02-06 | | Release date: | 2007-03-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | The structure of probable glutaminase from B. subtilis complexed with its inhibitor 6-diazo-5-oxo-L-norleucine
To be Published
|
|
1NNI
 
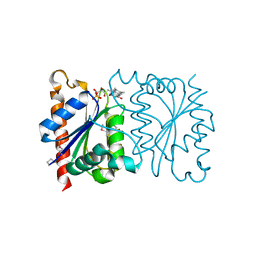 | | Azobenzene Reductase from Bacillus subtilis | | Descriptor: | FLAVIN MONONUCLEOTIDE, hypothetical protein yhda | | Authors: | Cuff, M.E, Kim, Y, Maj, L, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-13 | | Release date: | 2003-07-29 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Azobenzene Reductase from Bacillus subtilis
To be Published, 2003
|
|
1M3S
 
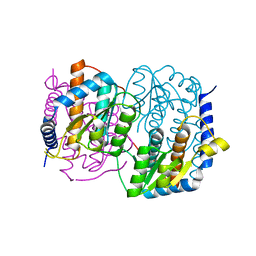 | | Crystal structure of YckF from Bacillus subtilis | | Descriptor: | Hypothetical protein yckf | | Authors: | Sanishvili, R, Wu, R, Kim, D.E, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-06-28 | | Release date: | 2003-01-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of Bacillus subtilis YckF: structural and functional evolution.
J.Struct.Biol., 148, 2004
|
|
1MK4
 
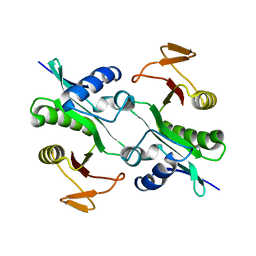 | | Structure of Protein of Unknown Function YqjY from Bacillus subtilis, Probable Acetyltransferase | | Descriptor: | Hypothetical protein yqjY | | Authors: | Zhang, R, Dementiva, I, Mo, A, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-08-28 | | Release date: | 2003-04-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7A crystal structure of a hypothetical protein yqjY
from Bacillus subtilis
To be Published
|
|
1NRW
 
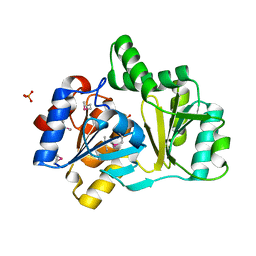 | | The structure of a HALOACID DEHALOGENASE-LIKE HYDROLASE FROM B. SUBTILIS | | Descriptor: | CALCIUM ION, PHOSPHATE ION, hypothetical protein, ... | | Authors: | Cuff, M.E, Kim, Y, Zhang, R, Joachimiak, A, Collart, F, Quartey, P, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-25 | | Release date: | 2003-07-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of a HALOACID DEHALOGENASE-LIKE HYDROLASE FROM B. SUBTILIS
To be Published
|
|
1PZX
 
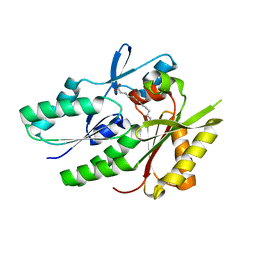 | | Hypothetical protein APC36103 from Bacillus stearothermophilus: a lipid binding protein | | Descriptor: | Hypothetical protein APC36103, PALMITIC ACID | | Authors: | Zhang, R, Osipiuk, J, Zhou, M, Alkire, R, Moy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-07-14 | | Release date: | 2004-01-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lipid binding protein APC36103 from Bacillus Stearothermophilus
To be Published
|
|
1NG6
 
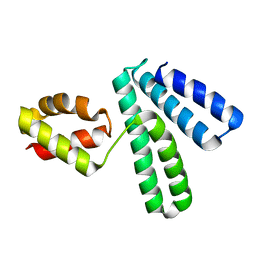 | | Structure of Cytosolic Protein of Unknown Function YqeY from Bacillus subtilis | | Descriptor: | Hypothetical protein yqeY | | Authors: | Zhang, R, Dementiva, I, Vinokour, E, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-12-16 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 1.4A crystal structure of hypothetical cytosolic protein
YQEY
To be Published
|
|
1NC5
 
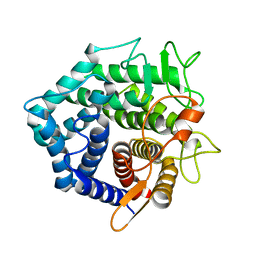 | | Structure of Protein of Unknown Function of YteR from Bacillus Subtilis | | Descriptor: | hypothetical protein yTER | | Authors: | Zhang, R, Lozondra, L, Korolev, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-12-04 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1.6 A crystal structure of YteR protein from Bacillus subtilis, a predicted lyase.
Proteins, 60, 2005
|
|
1NPY
 
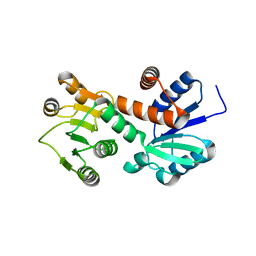 | | Structure of shikimate 5-dehydrogenase-like protein HI0607 | | Descriptor: | ACETYL GROUP, Hypothetical shikimate 5-dehydrogenase-like protein HI0607 | | Authors: | Korolev, S, Koroleva, O, Zarembinski, T, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-20 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of a Novel Shikimate Dehydrogenase from Haemophilus influenzae.
J.Biol.Chem., 280, 2005
|
|
3QWU
 
 | | Putative ATP-dependent DNA ligase from Aquifex aeolicus. | | Descriptor: | ADENOSINE, CALCIUM ION, DNA ligase, ... | | Authors: | Osipiuk, J, Quartey, P, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-28 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Putative ATP-dependent DNA ligase from Aquifex aeolicus.
To be Published
|
|
2FB5
 
 | | Structural Genomics; The crystal structure of the hypothetical membrane spanning protein from Bacillus cereus | | Descriptor: | hypothetical Membrane Spanning Protein | | Authors: | Zhang, R, Zhou, M, Ginell, S, Abdullah, J, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-08 | | Release date: | 2006-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The crystal structure of the hypothetical membrane spanning protein from Bacillus cereus
To be Published
|
|
2FB6
 
 | | Structure of Conserved Protein of Unknown Function BT1422 from Bacteroides thetaiotaomicron | | Descriptor: | conserved hypothetical protein | | Authors: | Osipiuk, J, Mulligan, R, Abdullah, J, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-08 | | Release date: | 2006-01-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | X-ray crystal structure of conserved hypothetical protein BT1422 from Bacteroides thetaiotaomicron
To be Published
|
|
2F9H
 
 | | The Crystal Structure of PTS System IIA Component from Enterococcus faecalis V583 | | Descriptor: | PTS system, IIA component | | Authors: | Kim, Y, Quartey, P, Moy, S, Bargassa, M, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-05 | | Release date: | 2006-01-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The Crystal Structure of PTS System IIA Component from Enterococcus faecalis V583
To be Published, 2006
|
|
