2KGT
 
 | |
2K25
 
 | |
2K22
 
 | |
2K24
 
 | |
7KNV
 
 | | Solution NMR structure of CDHR3 extracellular domain EC1 | | Descriptor: | CALCIUM ION, Cadherin-related family member 3 | | Authors: | Lee, W, Tonelli, M, Frederick, R.O, Watters, K.E, Markley, J.L, Palmenberg, A.C. | | Deposit date: | 2020-11-06 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Determination of the CDHR3 Rhinovirus-C Binding Domain, EC1
Viruses, 13, 2021
|
|
1PES
 
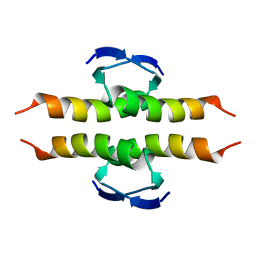 | | NMR SOLUTION STRUCTURE OF THE TETRAMERIC MINIMUM TRANSFORMING DOMAIN OF P53 | | Descriptor: | TUMOR SUPPRESSOR P53 | | Authors: | Lee, W, Harvey, T.S, Yin, Y, Yau, P, Litchfield, D, Arrowsmith, C.H. | | Deposit date: | 1994-11-24 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the tetrameric minimum transforming domain of p53.
Nat.Struct.Biol., 1, 1994
|
|
1PET
 
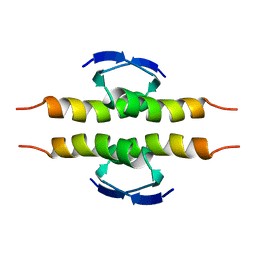 | | NMR SOLUTION STRUCTURE OF THE TETRAMERIC MINIMUM TRANSFORMING DOMAIN OF P53 | | Descriptor: | TUMOR SUPPRESSOR P53 | | Authors: | Lee, W, Harvey, T.S, Yin, Y, Yau, P, Litchfield, D, Arrowsmith, C.H. | | Deposit date: | 1994-11-24 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the tetrameric minimum transforming domain of p53.
Nat.Struct.Biol., 1, 1994
|
|
1VNA
 
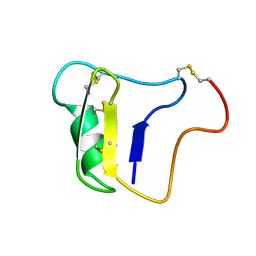 | |
1VNB
 
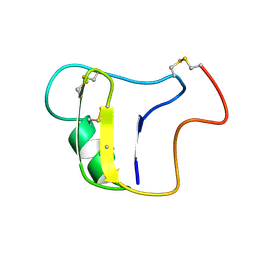 | |
2M5T
 
 | | Solution structure of the 2A proteinase from a common cold agent, human rhinovirus RV-C02, strain W12 | | Descriptor: | ZINC ION, human rhinovirus 2A proteinase | | Authors: | Lee, W, Frederick, R, Tonelli, M, Troupis, A.T, Reinin, N, Suchy, F.P, Moyer, K, Watters, K, Aceti, D, Palmenberg, A.C, Markley, J.L. | | Deposit date: | 2013-03-07 | | Release date: | 2014-03-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the 2A Protease from a Common Cold Agent, Human Rhinovirus C2, Strain W12.
Plos One, 9, 2014
|
|
2KX2
 
 | |
6NU4
 
 | | Solution structure of the Arabidopsis thaliana RALF8 peptide | | Descriptor: | Protein RALF-like 8 | | Authors: | Lee, W, Markley, J.L, Frederick, R.O, Miyoshi, H, Tonelli, M, Cornilescu, G, Cornilescu, C, Sussman, M.R. | | Deposit date: | 2019-01-30 | | Release date: | 2019-05-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Function and solution structure of the Arabidopsis thaliana RALF8 peptide.
Protein Sci., 28, 2019
|
|
7LU8
 
 | |
2M17
 
 | |
2ROH
 
 | | The DNA binding domain of RTBP1 | | Descriptor: | Telomere binding protein-1 | | Authors: | Lee, W, Ko, S. | | Deposit date: | 2008-03-22 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA binding domain of rice telomere binding protein RTBP1
Biochemistry, 48, 2009
|
|
1HVB
 
 | | CRYSTAL STRUCTURE OF STREPTOMYCES R61 DD-PEPTIDASE COMPLEXED WITH A NOVEL CEPHALOSPORIN ANALOG OF CELL WALL PEPTIDOGLYCAN | | Descriptor: | 5-{3-(S)-(4-(R)-ACETYLAMINO-4-CARBOXY-BUTYRYLAMINO)-3-[1-(R)-(1-(R)-CARBOXY-ETHYLCARBAMOYL)-ETHYLCARBAMOYL]-PROPYL}-2-( CARBOXY-PHENYLACETYLAMINO-METHYL)-3,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE | | Authors: | McDonough, M.A, Lee, W, Silvaggi, N.R, Mobashery, S, Kelly, J.A. | | Deposit date: | 2001-01-08 | | Release date: | 2001-02-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | A 1.2-A snapshot of the final step of bacterial cell wall biosynthesis.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
6KZ7
 
 | | The crystal structure of BAF155 SWIRM domain and N-terminal elongated hSNF5 RPT1 domain complex: Chromatin remodeling complex | | Descriptor: | SWI/SNF complex subunit SMARCC1, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1 | | Authors: | Lee, W, Han, J, Kim, I, Park, J.H, Joo, K, Lee, J, Suh, J.Y. | | Deposit date: | 2019-09-23 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A Coil-to-Helix Transition Serves as a Binding Motif for hSNF5 and BAF155 Interaction.
Int J Mol Sci, 21, 2020
|
|
6LZP
 
 | |
1R02
 
 | |
2ABY
 
 | | Solution structure of TA0743 from Thermoplasma acidophilum | | Descriptor: | hypothetical protein TA0743 | | Authors: | Kim, B, Jung, J, Hong, E, Yee, A, Arrowsmith, C.H, Lee, W. | | Deposit date: | 2005-07-18 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the conserved novel-fold protein TA0743 from Thermoplasma acidophilum.
Proteins, 62, 2006
|
|
6X6N
 
 | |
6O6W
 
 | | Solution structure of human myeloid-derived growth factor | | Descriptor: | Myeloid-derived growth factor | | Authors: | Bortnov, V, Tonelli, M, Lee, W, Markley, J.L, Mosher, D.F. | | Deposit date: | 2019-03-07 | | Release date: | 2019-11-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human myeloid-derived growth factor suggests a conserved function in the endoplasmic reticulum.
Nat Commun, 10, 2019
|
|
1RYJ
 
 | | Solution NMR Structure of Protein Mth1743 from Methanobacterium thermoautotrophicum. Ontario Centre for Structural Proteomics target MTH1743_1_70; Northeast Structural Genomics Consortium Target TT526. | | Descriptor: | unknown | | Authors: | Yee, A, Chang, X, Pineda-Lucena, A, Wu, B, Semesi, A, Le, B, Ramelot, T, Lee, G.M, Bhattacharyya, S, Gutierrez, P, Denisov, A, Lee, C.H, Cort, J.R, Kozlov, G, Liao, J, Finak, G, Chen, L, Wishart, D, Lee, W, McIntosh, L.P, Gehring, K, Kennedy, M.A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-12-22 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | AN NMR APPROACH TO STRUCTURAL PROTEOMICS
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1CQ0
 
 | | SOLUTION STRUCTURE OF A HUMAN HYPOCRETIN-2/OREXIN-B'SOLUTION STRUCTURE OF A HUMAN HYPOCRETIN-2/OREXIN-B ' | | Descriptor: | PROTEIN (NEW HYPOTHALAMIC NEUROPEPTIDE/OREXIN-B28) | | Authors: | Lee, K.-H, Bang, E.J, Chae, K.-J, Lee, D.W, Lee, W. | | Deposit date: | 1999-08-04 | | Release date: | 2000-01-10 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a new hypothalamic neuropeptide, human hypocretin-2/orexin-B.
Eur.J.Biochem., 266, 1999
|
|
1MNL
 
 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF A SWEET PROTEIN SINGLE-CHAIN MONELLIN (SCM) DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND DYNAMICAL SIMULATED ANNEALING CALCULATIONS, 21 STRUCTURES | | Descriptor: | MONELLIN | | Authors: | Lee, S.-Y, Lee, J.-H, Chang, H.-J, Jo, J.-M, Jung, J.-W, Lee, W. | | Deposit date: | 1998-08-06 | | Release date: | 1999-06-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a sweet protein single-chain monellin determined by nuclear magnetic resonance and dynamical simulated annealing calculations.
Biochemistry, 38, 1999
|
|
