5MX2
 
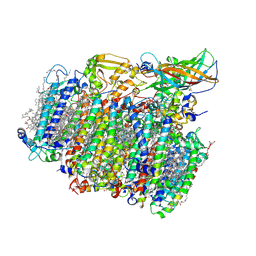 | | Photosystem II depleted of the Mn4CaO5 cluster at 2.55 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Zhang, M, Bommer, M, Chatterjee, R, Hussain, R, Kern, J, Yano, J, Dau, H, Dobbek, H, Zouni, A. | | Deposit date: | 2017-01-20 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Structural insights into the light-driven auto-assembly process of the water-oxidizing Mn4CaO5-cluster in photosystem II.
Elife, 6, 2017
|
|
2B1N
 
 | |
5XED
 
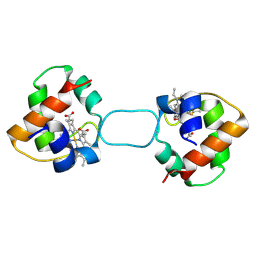 | | Heterodimer constructed from M61A PA cyt c551-HT cyt c552 and HT cyt c552-PA cyt c551 chimeric proteins | | Descriptor: | Cytochrome c-551,Cytochrome c-552, Cytochrome c-552,Cytochrome c-551, HEME C | | Authors: | Zhang, M, Nakanishi, T, Yamanaka, M, Nagao, S, Yanagisawa, S, Shomura, Y, Shibata, N, Ogura, T, Higuchi, Y, Hirota, S. | | Deposit date: | 2017-04-04 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Rational Design of Domain-Swapping-Based c-Type Cytochrome Heterodimers by Using Chimeric Proteins.
Chembiochem, 18, 2017
|
|
3L0B
 
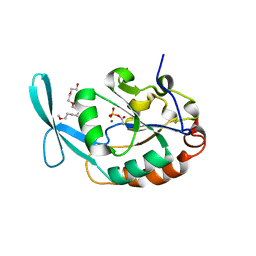 | | Crystal structure of SCP1 phosphatase D206A mutant phosphoryl-intermediate | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, Carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1, MAGNESIUM ION | | Authors: | Zhang, M, Zhang, Y. | | Deposit date: | 2009-12-09 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and functional analysis of the phosphoryl transfer reaction mediated by the human small C-terminal domain phosphatase, Scp1.
Protein Sci., 19, 2010
|
|
3L0Y
 
 | | Crystal structure OF SCP1 phosphatase D98A mutant | | Descriptor: | Carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1, MAGNESIUM ION | | Authors: | Zhang, M, Zhang, Y. | | Deposit date: | 2009-12-10 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional analysis of the phosphoryl transfer reaction mediated by the human small C-terminal domain phosphatase, Scp1.
Protein Sci., 19, 2010
|
|
3L0C
 
 | |
6IMA
 
 | | Crystal Structure of ALKBH1 without alpha-1 (N37-C369) | | Descriptor: | CITRIC ACID, Nucleic acid dioxygenase ALKBH1 | | Authors: | Zhang, M, Yang, S, Zhao, W, Li, H. | | Deposit date: | 2018-10-22 | | Release date: | 2020-01-22 | | Last modified: | 2020-04-08 | | Method: | X-RAY DIFFRACTION (2.593 Å) | | Cite: | Mammalian ALKBH1 serves as an N6-mA demethylase of unpairing DNA.
Cell Res., 30, 2020
|
|
4GEH
 
 | |
1PNT
 
 | |
4YVO
 
 | |
4YVQ
 
 | |
4QNH
 
 | | Calcium-calmodulin (T79D) complexed with the calmodulin binding domain from a small conductance potassium channel SK2-a | | Descriptor: | CALCIUM ION, Calmodulin, SULFATE ION, ... | | Authors: | Zhang, M, Pascal, J.M, Logothetis, D.E, Zhang, J.F. | | Deposit date: | 2014-06-17 | | Release date: | 2014-08-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Selective phosphorylation modulates the PIP2 sensitivity of the CaM-SK channel complex.
Nat.Chem.Biol., 10, 2014
|
|
4G28
 
 | | Calcium-calmodulin complexed with the calmodulin binding domain from a small conductance potassium channel splice variant and EBIO-1 | | Descriptor: | 1-ethyl-1,3-dihydro-2H-benzimidazol-2-one, CALCIUM ION, Calmodulin, ... | | Authors: | Zhang, M, Pascal, J.M, Zhang, J.-F. | | Deposit date: | 2012-07-11 | | Release date: | 2012-09-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Identification of the functional binding pocket for compounds targeting small-conductance Ca(2+)-activated potassium channels.
Nat Commun, 3, 2012
|
|
4G27
 
 | | Calcium-calmodulin complexed with the calmodulin binding domain from a small conductance potassium channel splice variant and phenylurea | | Descriptor: | 1-phenylurea, CALCIUM ION, Calmodulin, ... | | Authors: | Zhang, M, Pascal, J.M, Zhang, J.-F. | | Deposit date: | 2012-07-11 | | Release date: | 2012-09-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification of the functional binding pocket for compounds targeting small-conductance Ca(2+)-activated potassium channels.
Nat Commun, 3, 2012
|
|
1DG9
 
 | | CRYSTAL STRUCTURE OF BOVINE LOW MOLECULAR WEIGHT PTPASE COMPLEXED WITH HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, TYROSINE PHOSPHATASE | | Authors: | Zhang, M, Zhou, M, Van Etten, R.L, Stauffacher, C.V. | | Deposit date: | 1999-11-23 | | Release date: | 1999-12-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of bovine low molecular weight phosphotyrosyl phosphatase complexed with the transition state analog vanadate.
Biochemistry, 36, 1997
|
|
4RFS
 
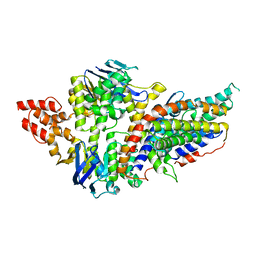 | | Structure of a pantothenate energy coupling factor transporter | | Descriptor: | Energy-coupling factor transporter ATP-binding protein EcfA1, Energy-coupling factor transporter ATP-binding protein EcfA2, Energy-coupling factor transporter transmembrane protein EcfT, ... | | Authors: | Zhang, M, Bao, Z, Zhao, Q, Guo, H, Xu, K, Zhang, P. | | Deposit date: | 2014-09-27 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.232 Å) | | Cite: | Structure of a pantothenate transporter and implications for ECF module sharing and energy coupling of group II ECF transporters.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1DMO
 
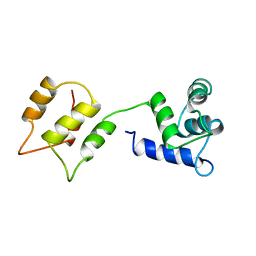 | | CALMODULIN, NMR, 30 STRUCTURES | | Descriptor: | CALMODULIN | | Authors: | Zhang, M, Tanaka, T, Ikura, M. | | Deposit date: | 1996-04-24 | | Release date: | 1996-08-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Calcium-induced conformational transition revealed by the solution structure of apo calmodulin.
Nat.Struct.Biol., 2, 1995
|
|
4J9Z
 
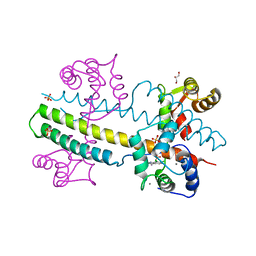 | | Calcium-calmodulin complexed with the calmodulin binding domain from a small conductance potassium channel splice variant and NS309 | | Descriptor: | (3E)-6,7-dichloro-3-(hydroxyimino)-1,3-dihydro-2H-indol-2-one, CALCIUM ION, Calmodulin, ... | | Authors: | Zhang, M, Pascal, J.M, Zhang, J.-F. | | Deposit date: | 2013-02-17 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Unstructured to structured transition of an intrinsically disordered protein peptide in coupling Ca2+-sensing and SK channel activation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4J9Y
 
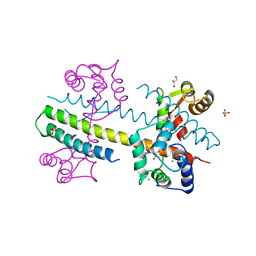 | | Calcium-calmodulin complexed with the calmodulin binding domain from a small conductance potassium channel splice variant | | Descriptor: | CALCIUM ION, Calmodulin, GLYCEROL, ... | | Authors: | Zhang, M, Pascal, J.M, Zhang, J.-F. | | Deposit date: | 2013-02-17 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Unstructured to structured transition of an intrinsically disordered protein peptide in coupling Ca2+-sensing and SK channel activation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1Z12
 
 | | Crystal Structure of Bovine Low Molecular Weight PTPase Complexed with Vanadate | | Descriptor: | Low molecular weight phosphotyrosine protein phosphatase, VANADATE ION | | Authors: | Zhang, M, Zhou, M, Van Etten, R.L, Stauffacher, C.V. | | Deposit date: | 2005-03-03 | | Release date: | 2005-04-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Bovine Low Molecular Weight Phosphotyrosyl Phosphatase Complexed with the Transition State Analog Vanadate
Biochemistry, 36, 1997
|
|
1Z13
 
 | | Crystal Structure of Bovine Low Molecular Weight PTPase Complexed with Molybdate | | Descriptor: | Low molecular weight phosphotyrosine protein phosphatase, MOLYBDATE ION | | Authors: | Zhang, M, Zhou, M, Van Etten, R.L, Stauffacher, C.V. | | Deposit date: | 2005-03-03 | | Release date: | 2005-04-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Bovine Low Molecular Weight Phosphotyrosyl Phosphatase Complexed with the Transition State Analog Vanadate
Biochemistry, 36, 1997
|
|
3PGL
 
 | | Crystal structure of human small C-terminal domain phosphatase 1 (Scp1) bound to rabeprazole | | Descriptor: | 2-[(R)-{[4-(3-methoxypropoxy)-3-methylpyridin-2-yl]methyl}sulfinyl]-1H-benzimidazole, Carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1, MAGNESIUM ION | | Authors: | Zhang, M, Cho, E.J, Burstein, G, Siegel, D, Zhang, Y. | | Deposit date: | 2010-11-02 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Selective inactivation of a human neuronal silencing phosphatase by a small molecule inhibitor.
Acs Chem.Biol., 6, 2011
|
|
6L5R
 
 | | crystal structure of GgCGT in complex with UDP-Glu | | Descriptor: | 3-(4-HYDROXYPHENYL)-1-(2,4,6-TRIHYDROXYPHENYL)PROPAN-1-ONE, GgCGT, URIDINE-5'-DIPHOSPHATE | | Authors: | Zhang, M, Li, F.D, Ye, M. | | Deposit date: | 2019-10-24 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Functional Characterization and Structural Basis of an Efficient Di-C-glycosyltransferase fromGlycyrrhiza glabra.
J.Am.Chem.Soc., 142, 2020
|
|
6L5P
 
 | | crystal structure of GgCGT in complex with UDP-Glu | | Descriptor: | GgCGT, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Zhang, M, Li, F.D, Ye, M. | | Deposit date: | 2019-10-24 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Functional Characterization and Structural Basis of an Efficient Di-C-glycosyltransferase fromGlycyrrhiza glabra.
J.Am.Chem.Soc., 142, 2020
|
|
6L5S
 
 | | crystal structure of GgCGT in complex with UDP-Glu | | Descriptor: | 3-(4-HYDROXYPHENYL)-1-(2,4,6-TRIHYDROXYPHENYL)PROPAN-1-ONE, GLYCEROL, GgCGT, ... | | Authors: | Zhang, M, Li, F.D, Ye, M. | | Deposit date: | 2019-10-24 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.914 Å) | | Cite: | Functional Characterization and Structural Basis of an Efficient Di-C-glycosyltransferase fromGlycyrrhiza glabra.
J.Am.Chem.Soc., 142, 2020
|
|
