6IDE
 
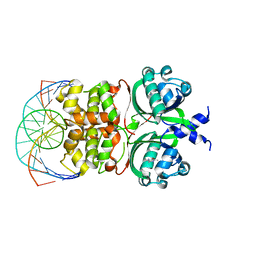 | | Crystal structure of the Vibrio cholera VqmA-Ligand-DNA complex provides molecular mechanisms for drug design | | Descriptor: | 3,5-dimethylpyrazin-2-ol, DNA (5'-D(*AP*GP*GP*GP*GP*GP*GP*AP*AP*AP*TP*CP*CP*CP*CP*CP*CP*T)-3'), DNA (5'-D(*AP*GP*GP*GP*GP*GP*GP*AP*TP*TP*TP*CP*CP*CP*CP*CP*CP*T)-3'), ... | | Authors: | Wu, H, Li, M.J, Guo, H.J, Zhou, H, Li, B, Xu, Q, Xu, C.Y, Yu, F, He, J.H. | | Deposit date: | 2018-09-09 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of theVibrio choleraeVqmA-ligand-DNA complex provides insight into ligand-binding mechanisms relevant for drug design.
J. Biol. Chem., 294, 2019
|
|
2CAS
 
 | |
1CDJ
 
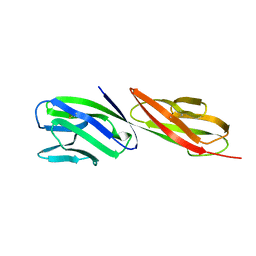 | | STRUCTURE OF T-CELL SURFACE GLYCOPROTEIN CD4 | | Descriptor: | T-CELL SURFACE GLYCOPROTEIN CD4 | | Authors: | Wu, H, Myszka, D, Tendian, S.W, Brouillette, C.G, Sweet, R.W, Chaiken, I.M, Hendrickson, W.A. | | Deposit date: | 1996-11-11 | | Release date: | 1997-04-01 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Kinetic and structural analysis of mutant CD4 receptors that are defective in HIV gp120 binding.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1CDU
 
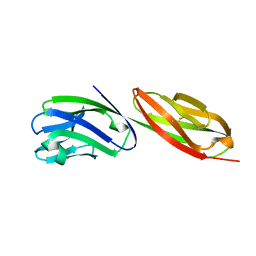 | | STRUCTURE OF T-CELL SURFACE GLYCOPROTEIN CD4 MUTANT WITH PHE 43 REPLACED BY VAL | | Descriptor: | T-CELL SURFACE GLYCOPROTEIN CD4 | | Authors: | Wu, H, Myszka, D, Tendian, S.W, Brouillette, C.G, Sweet, R.W, Chaiken, I.M, Hendrickson, W.A. | | Deposit date: | 1996-11-11 | | Release date: | 1997-04-01 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Kinetic and structural analysis of mutant CD4 receptors that are defective in HIV gp120 binding.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1CDY
 
 | | STRUCTURE OF T-CELL SURFACE GLYCOPROTEIN CD4 MUTANT WITH GLY 47 REPLACED BY SER | | Descriptor: | T-CELL SURFACE GLYCOPROTEIN CD4 | | Authors: | Wu, H, Myszka, D, Tendian, S.W, Brouillette, C.G, Sweet, R.W, Chaiken, I.M, Hendrickson, W.A. | | Deposit date: | 1996-11-11 | | Release date: | 1997-04-01 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and structural analysis of mutant CD4 receptors that are defective in HIV gp120 binding.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
6R2M
 
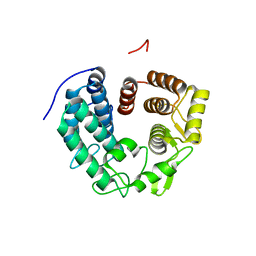 | | Crystal structure of PssZ from Listeria monocytogenes | | Descriptor: | Glycoside transferase | | Authors: | Wu, H, Cheng, J, Qiao, S, Li, D, Ma, L. | | Deposit date: | 2019-03-18 | | Release date: | 2019-07-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.617 Å) | | Cite: | Crystal structure of the glycoside hydrolase PssZ from Listeria monocytogenes.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
1HCN
 
 | | STRUCTURE OF HUMAN CHORIONIC GONADOTROPIN AT 2.6 ANGSTROMS RESOLUTION FROM MAD ANALYSIS OF THE SELENOMETHIONYL PROTEIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HUMAN CHORIONIC GONADOTROPIN | | Authors: | Wu, H, Lustbader, J.W, Liu, Y, Canfield, R.E, Hendrickson, W.A. | | Deposit date: | 1994-07-01 | | Release date: | 1994-09-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of human chorionic gonadotropin at 2.6 A resolution from MAD analysis of the selenomethionyl protein.
Structure, 2, 1994
|
|
2QTH
 
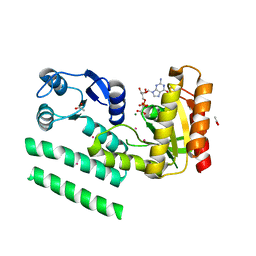 | | Crystal structure of a GTP-binding protein from the hyperthermophilic archaeon Sulfolobus solfataricus in complex with GDP | | Descriptor: | ACETATE ION, CADMIUM ION, GTP-binding protein, ... | | Authors: | Wu, H, Sun, L, Brouns, S.J, Fu, S, Rao, Z, Van der Oost, J. | | Deposit date: | 2007-08-02 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a GTP-binding protein from the hyperthermophilic archaeon Sulfolobus solfataricus.
To be Published
|
|
2QTF
 
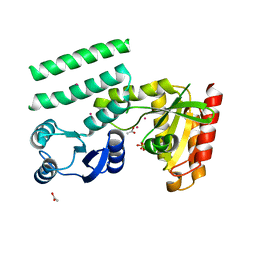 | | Crystal structure of a GTP-binding protein from the hyperthermophilic archaeon Sulfolobus solfataricus | | Descriptor: | ACETATE ION, CADMIUM ION, GTP-binding protein, ... | | Authors: | Wu, H, Sun, L, Brouns, S.J, Fu, S, Rao, Z, Van der Oost, J. | | Deposit date: | 2007-08-02 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a GTP-binding protein from the hyperthermophilic archaeon Sulfolobus solfataricus.
To be Published
|
|
2FJ4
 
 | |
5UNA
 
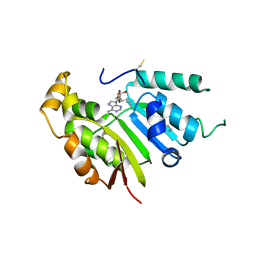 | | Fragment of 7SK snRNA methylphosphate capping enzyme | | Descriptor: | 7SK snRNA methylphosphate capping enzyme, S-ADENOSYL-L-HOMOCYSTEINE, unidentified peptide section/fragment | | Authors: | Wu, H, Tempel, W, Dombrovski, L, McCarthy, A.A, Loppnau, P, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-01-30 | | Release date: | 2017-03-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Fragment of 7SK snRNA methylphosphate capping enzyme
To Be Published
|
|
1K4B
 
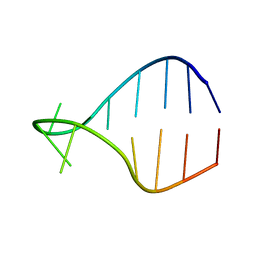 | | STRUCTURE OF AGUU RNA TETRALOOP, NMR, 20 STRUCTURES | | Descriptor: | 5'-R(*GP*GP*UP*UP*CP*AP*GP*UP*UP*GP*AP*AP*CP*C)-3' | | Authors: | Wu, H, Yang, P.K, Butcher, S.E, Kang, S, Chanfreau, G, Feigon, J. | | Deposit date: | 2001-10-07 | | Release date: | 2001-12-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A novel family of RNA tetraloop structure forms the recognition site for Saccharomyces cerevisiae RNase III.
EMBO J., 20, 2001
|
|
1K4A
 
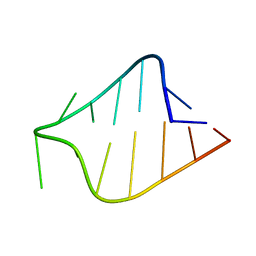 | | STRUCTURE OF AGAA RNA TETRALOOP, NMR, 20 STRUCTURES | | Descriptor: | 5'-R(*GP*GP*UP*UP*CP*AP*GP*AP*AP*GP*AP*AP*CP*C)-3' | | Authors: | Wu, H, Yang, P.K, Butcher, S.E, Kang, S, Chanfreau, G, Feigon, J. | | Deposit date: | 2001-10-07 | | Release date: | 2001-12-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A novel family of RNA tetraloop structure forms the recognition site for Saccharomyces cerevisiae RNase III.
EMBO J., 20, 2001
|
|
8EUF
 
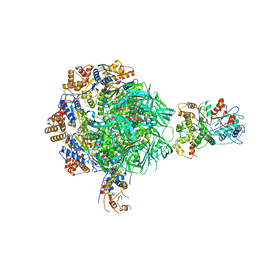 | | Class2 of the INO80-Nucleosome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling ATPase INO80, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Narlikar, G, Cheng, Y.F. | | Deposit date: | 2022-10-18 | | Release date: | 2023-07-12 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8EUJ
 
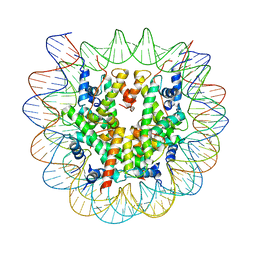 | | Class2 of the INO80-Nucleosome complex | | Descriptor: | DNA (147-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Narlikar, G, Cheng, Y.F. | | Deposit date: | 2022-10-18 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8ETT
 
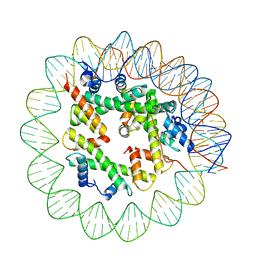 | | Class1 of the INO80-Hexasome complex | | Descriptor: | DNA (110-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Cheng, Y.F, Narlikar, G. | | Deposit date: | 2022-10-17 | | Release date: | 2023-07-12 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (6.68 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8ETV
 
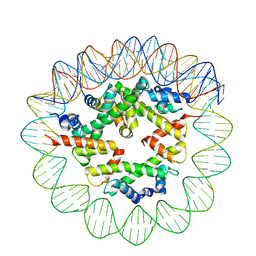 | | Class2 of the INO80-Hexasome complex | | Descriptor: | DNA (110-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Cheng, Y.F, Narlikar, G. | | Deposit date: | 2022-10-17 | | Release date: | 2023-07-12 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8EU2
 
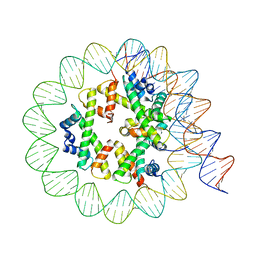 | | Class3 of the INO80-Hexasome complex | | Descriptor: | DNA (110-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Cheng, Y.F, Narlikar, G. | | Deposit date: | 2022-10-18 | | Release date: | 2023-07-12 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8EUE
 
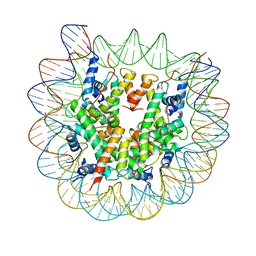 | | Class1 of the INO80-Nucleosome complex | | Descriptor: | DNA (147-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Narlikar, G, Cheng, Y.F. | | Deposit date: | 2022-10-18 | | Release date: | 2023-07-12 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8EU9
 
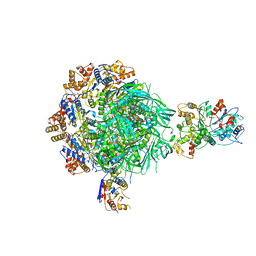 | | Class1 of the INO80-Nucleosome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling ATPase INO80, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Cheng, Y.F, Narlikar, G. | | Deposit date: | 2022-10-18 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8ETS
 
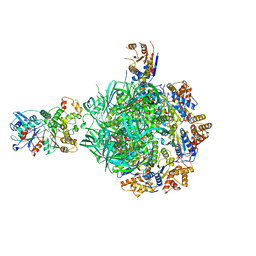 | | Class1 of the INO80-Hexasome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling ATPase INO80, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Cheng, Y.F, Narlikar, G. | | Deposit date: | 2022-10-17 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8ETU
 
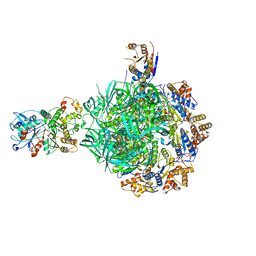 | | Class2 of the INO80-Hexasome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling ATPase INO80, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Cheng, Y.F, Narlikar, G. | | Deposit date: | 2022-10-17 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8ETW
 
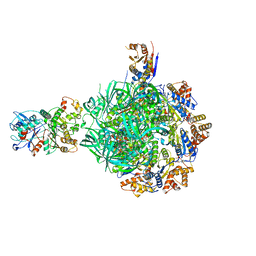 | | Class3 of INO80-Hexasome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling ATPase INO80, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Narlikar, G, Cheng, Y.F. | | Deposit date: | 2022-10-17 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
2P0W
 
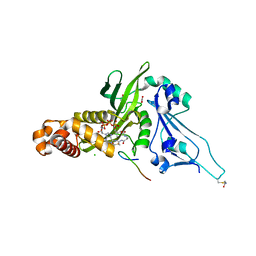 | | Human histone acetyltransferase 1 (HAT1) | | Descriptor: | ACETAMIDE, ACETATE ION, ACETYL COENZYME *A, ... | | Authors: | Wu, H, Min, J, Zeng, H, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-03-01 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of human histone acetyltransferase 1 (HAT1) in complex with acetylcoenzyme A and histone peptide H4
To be Published
|
|
1T4L
 
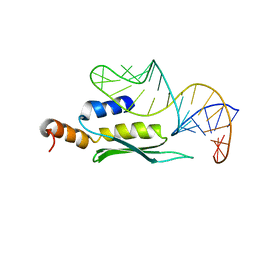 | | Solution structure of double-stranded RNA binding domain of S. cerevisiae RNase III (Rnt1p) in complex with the 5' terminal RNA hairpin of snR47 precursor | | Descriptor: | 5' terminal hairpin of snR47 precursor, Ribonuclease III | | Authors: | Wu, H, Henras, A, Chanfreau, G, Feigon, J. | | Deposit date: | 2004-04-29 | | Release date: | 2004-06-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recognition of the AGNN tetraloop RNA fold by the double-stranded RNA-binding domain of Rnt1p RNase III.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
