6VJ2
 
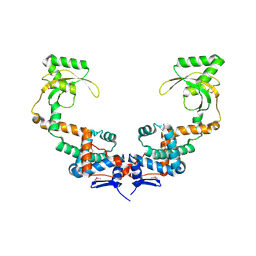 | | 3.10 Angstrom Resolution Crystal Structure of Foldase Protein (PrsA) from Lactococcus lactis | | Descriptor: | Foldase | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-01-14 | | Release date: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | 3.10 Angstrom Resolution Crystal Structure of Foldase Protein (PrsA) from Lactococcus lactis
To Be Published
|
|
6VJ4
 
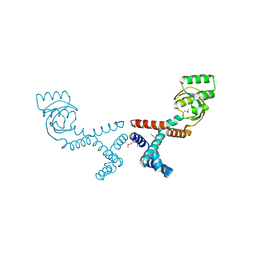 | | 1.70 Angstrom Resolution Crystal Structure of Peptidylprolyl Isomerase (PrsA) from Bacillus anthracis | | Descriptor: | Peptidylprolyl isomerase PrsA | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Wiersum, G, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-01-14 | | Release date: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.70 Angstrom Resolution Crystal Structure of Peptidylprolyl Isomerase (PrsA) from Bacillus anthracis
To Be Published
|
|
4YGO
 
 | | Dodecameric structure of spermidine N-acetyltransferase from Vibrio cholerae in intermediate state | | Descriptor: | CALCIUM ION, METHANOL, Spermidine n1-acetyltransferase | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-02-26 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate-Induced Allosteric Change in the Quaternary Structure of the Spermidine N-Acetyltransferase SpeG.
J.Mol.Biol., 427, 2015
|
|
3R2I
 
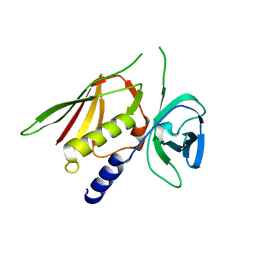 | | Crystal Structure of Superantigen-like Protein, Exotoxin SACOL0473 from Staphylococcus aureus subsp. aureus COL | | Descriptor: | Exotoxin 5 | | Authors: | Filippova, E.V, Minasov, G, Halavaty, A, Shuvalova, L, Winsor, J, Dubrovska, I, Kiryukhina, O, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-14 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Superantigen-like Protein, Exotoxin SACOL0473 from Staphylococcus aureus subsp. aureus COL
TO BE PUBLISHED
|
|
6MUQ
 
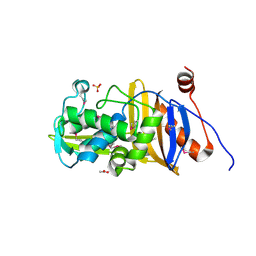 | | 1.67 Angstrom Resolution Crystal Structure of Murein-DD-endopeptidase from Yersinia enterocolitica. | | Descriptor: | ACETATE ION, Murein-DD-endopeptidase, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-23 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | 1.67 Angstrom Resolution Crystal Structure of Murein-DD-endopeptidase from Yersinia enterocolitica.
To Be Published
|
|
7JZ0
 
 | | Crystal Structure of SARS-CoV-2 Nsp16/10 Heterodimer in Complex with (m7GpppA2m)pUpUpApApA (Cap-1) and S-Adenosyl-L-homocysteine (SAH). | | Descriptor: | 2'-O-methyltransferase, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of SARS-CoV-2 2'-O-methyltransferase heterodimer with RNA Cap analog and sulfates bound reveals new strategies for structure-based inhibitor design
Biorxiv, 2020
|
|
6PNV
 
 | | 1.42 Angstrom Resolution Crystal Structure of Translocation Protein TolB from Salmonella enterica | | Descriptor: | POTASSIUM ION, SODIUM ION, Tol-Pal system protein TolB | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-07-03 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | 1.42 Angstrom Resolution Crystal Structure of Translocation Protein TolB from Salmonella enterica
To Be Published
|
|
3F0A
 
 | | Structure of a putative n-acetyltransferase (ta0374) in complex with acetyl-coa from thermoplasma acidophilum | | Descriptor: | ACETYL COENZYME *A, CHLORIDE ION, N-ACETYLTRANSFERASE, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-24 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the novel PaiA N-acetyltransferase from Thermoplasma acidophilum involved in the negative control of sporulation and degradative enzyme production.
Proteins, 79, 2011
|
|
3FIX
 
 | | Crystal structure of a putative n-acetyltransferase (ta0374) from thermoplasma acidophilum | | Descriptor: | 1,2-ETHANEDIOL, N-ACETYLTRANSFERASE | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-12 | | Release date: | 2009-01-13 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the novel PaiA N-acetyltransferase from Thermoplasma acidophilum involved in the negative control of sporulation and degradative enzyme production.
Proteins, 79, 2011
|
|
2I0Z
 
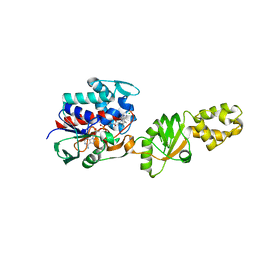 | | Crystal structure of a FAD binding protein from Bacillus cereus, a putative NAD(FAD)-utilizing dehydrogenases | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, NAD(FAD)-utilizing dehydrogenases | | Authors: | Minasov, G, Shuvalova, L, Vorontsov, I.I, Kiryukhina, O, Abdullah, J, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-11 | | Release date: | 2006-08-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of a FAD binding protein from Bacillus cereus, a putative NAD(FAD)-utilizing dehydrogenases
To be Published
|
|
3LZ8
 
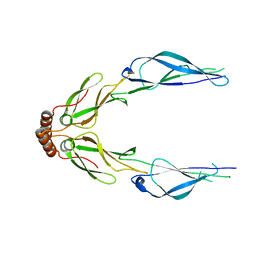 | | Structure of a putative chaperone dnaj from klebsiella pneumoniae subsp. pneumoniae mgh 78578 at 2.9 a resolution. | | Descriptor: | Putative chaperone DnaJ | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Bearden, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-01 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a Putative Chaperone Dnaj from Klebsiella Pneumoniae Subsp. Pneumoniae Mgh 78578 at 2.9 A Resolution.
To be Published
|
|
3M6Y
 
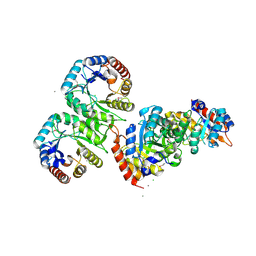 | | Structure of 4-hydroxy-2-oxoglutarate aldolase from bacillus cereus at 1.45 a resolution. | | Descriptor: | 4-Hydroxy-2-oxoglutarate aldolase, CALCIUM ION, CHLORIDE ION | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, F.W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-16 | | Release date: | 2010-04-07 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of 4-Hydroxy-2-Oxoglutarate Aldolase from Bacillus Cereus at 1.45 A Resolution.
To be Published
|
|
3MAH
 
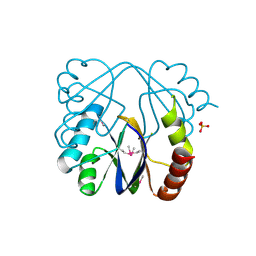 | | A putative c-terminal regulatory domain of aspartate kinase from porphyromonas gingivalis w83. | | Descriptor: | Aspartokinase, SULFATE ION | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Moy, S, Joachimiak, A, Anderson, F.W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-23 | | Release date: | 2010-04-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A Putative C-Terminal Regulatory Domain of Aspartate Kinase from Porphyromonas Gingivalis W83.
To be Published
|
|
3MVN
 
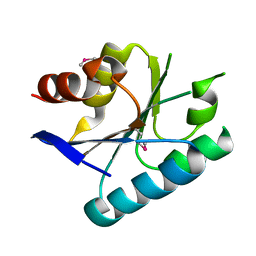 | | Crystal structure of a domain from a putative UDP-N-acetylmuramate:L-alanyl-gamma-D-glutamyl-medo-diaminopimelate ligase from Haemophilus ducreyi 35000HP | | Descriptor: | UDP-N-acetylmuramate:L-alanyl-gamma-D-glutamyl-medo-diaminopimelate ligase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, F.W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-04 | | Release date: | 2010-06-16 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a domain from a putative UDP-N-acetylmuramate:L-alanyl-gamma-D-glutamyl-medo-diaminopimelate ligase from Haemophilus ducreyi 35000HP
To be Published
|
|
6AWA
 
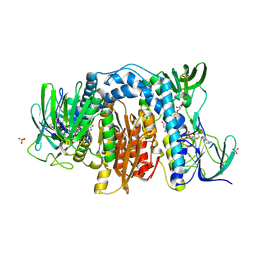 | | 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Pseudomonas putida in Complex with FAD and Adenosine-5'-monophosphate. | | Descriptor: | ADENOSINE MONOPHOSPHATE, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-09-05 | | Release date: | 2017-10-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Pseudomonas putida in Complex with FAD and Adenosine-5'-monophosphate.
To Be Published
|
|
6AZI
 
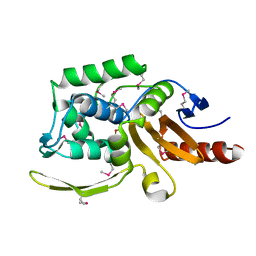 | | 1.75 Angstrom Resolution Crystal Structure of D-alanyl-D-alanine Endopeptidase from Enterobacter cloacae in Complex with Covalently Bound Boronic Acid | | Descriptor: | BORATE ION, D-alanyl-D-alanine endopeptidase | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-09-11 | | Release date: | 2017-10-04 | | Last modified: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | 1.75 Angstrom Resolution Crystal Structure of D-alanyl-D-alanine Endopeptidase from Enterobacter cloacae in Complex with Covalently Bound Boronic Acid.
To be Published
|
|
6B8D
 
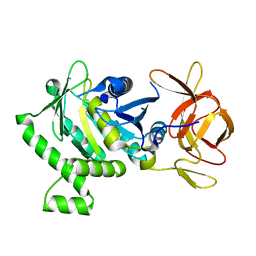 | | 1.78 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-405) of Elongation Factor G from Haemophilus influenzae | | Descriptor: | CHLORIDE ION, Elongation factor G | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-10-06 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | 1.78 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-405) of Elongation Factor G from Haemophilus influenzae.
To Be Published
|
|
6BLB
 
 | | 1.88 Angstrom Resolution Crystal Structure Holliday Junction ATP-dependent DNA Helicase (RuvB) from Pseudomonas aeruginosa in Complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvB, TRIETHYLENE GLYCOL | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-09 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | 1.88 Angstrom Resolution Crystal Structure Holliday Junction ATP-dependent DNA Helicase (RuvB) from Pseudomonas aeruginosa in Complex with ADP.
To be Published
|
|
7SUA
 
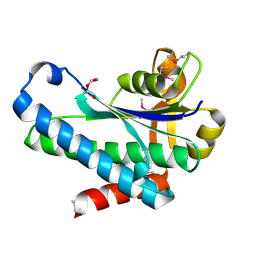 | | Crystal Structure of the Hypothetical Protein (ACX60_00475) from Acinetobacter baumannii | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DUF4175 domain-containing protein | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-30 | | Last modified: | 2023-02-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of the Hypothetical Protein (ACX60_00475) from Acinetobacter baumannii
To be Published
|
|
7TZP
 
 | | Crystal Structure of Putataive Short-Chain Dehydrogenase/Reductase (FabG) from Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044 in Complex with NADH | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3-oxoacyl-ACP reductase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-02-16 | | Release date: | 2022-03-02 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
8UFM
 
 | | Crystal Structure of L516C/Y647C Mutant of SARS-Unique Domain (SUD) from SARS-CoV-2 | | Descriptor: | ACETATE ION, FORMIC ACID, Papain-like protease nsp3, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-10-04 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of L516C/Y647C Mutant of SARS-Unique Domain (SUD) from SARS-CoV-2
To Be Published
|
|
8UFL
 
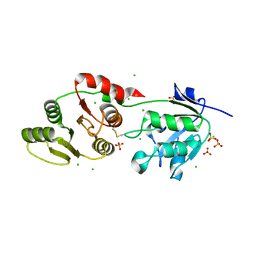 | | Crystal Structure of SARS-Unique Domain (SUD) of Nsp3 from SARS coronavirus | | Descriptor: | CHLORIDE ION, Papain-like protease nsp3, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-10-04 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal Structure of SARS-Unique Domain (SUD) of Nsp3 from SARS coronavirus
To Be Published
|
|
7ULT
 
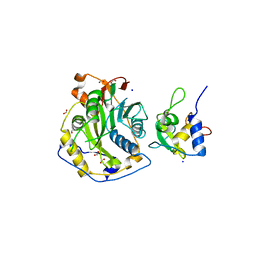 | | Crystal Structure of SARS-CoV-2 Nsp16/10 Heterodimer Apo-Form. | | Descriptor: | 2'-O-methyltransferase, FORMIC ACID, Non-structural protein 10, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-04-05 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of SARS-CoV-2 Nsp16/10 Heterodimer Apo-Form.
To Be Published
|
|
4YF1
 
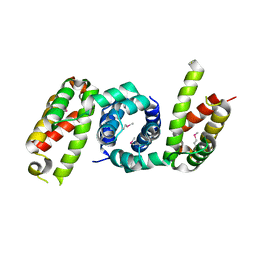 | | 1.85 angstrom crystal structure of lmo0812 from Listeria monocytogenes EGD-e | | Descriptor: | CITRATE ANION, Lmo0812 protein, SODIUM ION | | Authors: | Krishna, S.N, Light, S.H, Filippova, E.V, Minasov, G, Kiryukhina, O, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-02-24 | | Release date: | 2015-03-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 angstrom crystal structure of lmo0812 from Listeria monocytogenes EGD-e
To Be Published
|
|
4Z8Z
 
 | | Crystal structure of the hypothetical protein from Ruminiclostridium thermocellum ATCC 27405 | | Descriptor: | Uncharacterized protein | | Authors: | Filippova, E.V, Wawrzak, Z, Kiryukhina, O, Endres, M, Joachimiak, J, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-04-09 | | Release date: | 2015-05-06 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the hypothetical protein from Ruminiclostridium thermocellum ATCC 27405
To Be Published
|
|
