7F0X
 
 | | A SARS-CoV-2 neutralizing antibody | | Descriptor: | Antibody, Spike protein S1 | | Authors: | Zhang, G, Li, X, Guo, Y, Wang, Y, Yuan, S. | | Deposit date: | 2021-06-07 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A SARS-CoV-2 neutralizing
antibody
To Be Published
|
|
7F12
 
 | | A SARS-CoV-2 neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody, Spike protein S1 | | Authors: | Zhang, G, Li, X, Guo, Y, Wang, Y, Yuan, S. | | Deposit date: | 2021-06-07 | | Release date: | 2022-06-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A SARS-CoV-2 neutralizing
antibody
To Be Published
|
|
7F15
 
 | | A SARS-CoV-2 neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody, Spike protein S1 | | Authors: | Zhang, G, Li, X, Guo, Y, Wang, Y, Yuan, S. | | Deposit date: | 2021-06-07 | | Release date: | 2022-06-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A SARS-CoV-2 neutralizing
antibody
To Be Published
|
|
6NT5
 
 | | Cryo-EM structure of full-length human STING in the apo state | | Descriptor: | Stimulator of interferon protein | | Authors: | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | Deposit date: | 2019-01-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP.
Nature, 567, 2019
|
|
6NT7
 
 | | Cryo-EM structure of full-length chicken STING in the cGAMP-bound dimeric state | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | Deposit date: | 2019-01-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP.
Nature, 567, 2019
|
|
6NT8
 
 | | Cryo-EM structure of full-length chicken STING in the cGAMP-bound tetrameric state | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | Deposit date: | 2019-01-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP.
Nature, 567, 2019
|
|
6NT6
 
 | | Cryo-EM structure of full-length chicken STING in the apo state | | Descriptor: | Stimulator of interferon genes protein | | Authors: | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | Deposit date: | 2019-01-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP.
Nature, 567, 2019
|
|
4EQA
 
 | | Crystal structure of PA1844 in complex with PA1845 from Pseudomonas aeruginosa PAO1 | | Descriptor: | Putative uncharacterized protein | | Authors: | Shang, G, Li, N, Zhang, J, Lu, D, Yu, Q, Zhao, Y, Liu, X, Xu, S, Gu, L. | | Deposit date: | 2012-04-18 | | Release date: | 2012-09-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insight into how Pseudomonas aeruginosa peptidoglycanhydrolase Tse1 and its immunity protein Tsi1 function.
Biochem.J., 448, 2012
|
|
5V6H
 
 | |
5V6E
 
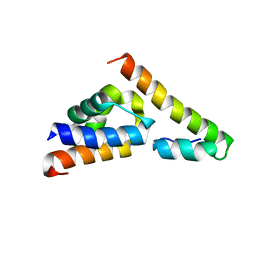 | |
5V6T
 
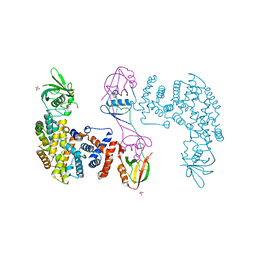 | | The Plexin D1 intracellular region in complex with GIPC1 | | Descriptor: | PDZ domain-containing protein GIPC1, Plexin-D1, SULFATE ION | | Authors: | Shang, G, Zhang, X. | | Deposit date: | 2017-03-17 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.189 Å) | | Cite: | Structure analyses reveal a regulated oligomerization mechanism of the PlexinD1/GIPC/myosin VI complex.
Elife, 6, 2017
|
|
5V6B
 
 | | Crystal structure of GIPC1 | | Descriptor: | PDZ domain-containing protein GIPC1 | | Authors: | Shang, G, Zhang, X. | | Deposit date: | 2017-03-16 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure analyses reveal a regulated oligomerization mechanism of the PlexinD1/GIPC/myosin VI complex.
Elife, 6, 2017
|
|
5V6R
 
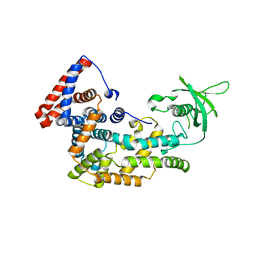 | | Structure of Plexin D1 intracellular domain | | Descriptor: | Plexin-D1 | | Authors: | Shang, G, Zhang, X. | | Deposit date: | 2017-03-17 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure analyses reveal a regulated oligomerization mechanism of the PlexinD1/GIPC/myosin VI complex.
Elife, 6, 2017
|
|
4EQ8
 
 | | Crystal structure of PA1844 from Pseudomonas aeruginosa PAO1 | | Descriptor: | GLYCEROL, Putative uncharacterized protein | | Authors: | Shang, G, Li, N, Zhang, J, Lu, D, Yu, Q, Zhao, Y, Liu, X, Xu, S, Gu, L. | | Deposit date: | 2012-04-18 | | Release date: | 2012-09-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.392 Å) | | Cite: | Structural insight into how Pseudomonas aeruginosa peptidoglycanhydrolase Tse1 and its immunity protein Tsi1 function.
Biochem.J., 448, 2012
|
|
1AB8
 
 | |
1BDF
 
 | |
2LSP
 
 | | solution structures of BRD4 second bromodomain with NF-kB-K310ac peptide | | Descriptor: | Bromodomain-containing protein 4, NF-kB-K310ac peptide | | Authors: | Zhang, G, Liu, R, Zhong, Y, Plotnikov, A.N, Zhang, W, Rusinova, E, Gerona-Nevarro, G, Moshkina, N, Joshua, J, Chuang, P.Y, Ohlmeyer, M, He, J, Zhou, M.-M. | | Deposit date: | 2012-05-03 | | Release date: | 2012-07-18 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Down-regulation of NF-kappa B transcriptional activity in HIV-associated kidney disease by BRD4 inhibition.
J.Biol.Chem., 287, 2012
|
|
1KJ1
 
 | | MANNOSE-SPECIFIC AGGLUTININ (LECTIN) FROM GARLIC (ALLIUM SATIVUM) BULBS COMPLEXED WITH ALPHA-D-MANNOSE | | Descriptor: | alpha-D-mannopyranose, lectin I, lectin II | | Authors: | Ramachandraiah, G, Chandra, N.R, Surolia, A, Vijayan, M. | | Deposit date: | 2001-12-04 | | Release date: | 2002-02-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Re-refinement using reprocessed data to improve the quality of the structure: a case study involving garlic lectin.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1SWV
 
 | | Crystal structure of the D12A mutant of phosphonoacetaldehyde hydrolase complexed with magnesium | | Descriptor: | MAGNESIUM ION, phosphonoacetaldehyde hydrolase | | Authors: | Zhang, G, Morais, M.C, Dai, J, Zhang, W, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2004-03-30 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Investigation of metal ion binding in phosphonoacetaldehyde hydrolase identifies sequence markers for metal-activated enzymes of the HAD enzyme superfamily
Biochemistry, 43, 2004
|
|
1SWW
 
 | | Crystal structure of the phosphonoacetaldehyde hydrolase D12A mutant complexed with magnesium and substrate phosphonoacetaldehyde | | Descriptor: | MAGNESIUM ION, PHOSPHONOACETALDEHYDE, phosphonoacetaldehyde hydrolase | | Authors: | Zhang, G, Morais, M.C, Dai, J, Zhang, W, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2004-03-30 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Investigation of metal ion binding in phosphonoacetaldehyde hydrolase identifies sequence markers for metal-activated enzymes of the HAD enzyme superfamily
Biochemistry, 43, 2004
|
|
8HBB
 
 | |
8HB2
 
 | |
8HAZ
 
 | |
1ZOL
 
 | | native beta-PGM | | Descriptor: | MAGNESIUM ION, beta-phosphoglucomutase | | Authors: | Zhang, G, Tremblay, L.W, Dai, J, Wang, L, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2005-05-13 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytic cycling in beta-phosphoglucomutase: a kinetic and structural analysis
Biochemistry, 44, 2005
|
|
3RFZ
 
 | | Crystal structure of the FimD usher bound to its cognate FimC:FimH substrate | | Descriptor: | Chaperone protein fimC, Outer membrane usher protein, type 1 fimbrial synthesis, ... | | Authors: | Phan, G, Remaut, H, Lebedev, A, Geibel, S, Waksman, G. | | Deposit date: | 2011-04-07 | | Release date: | 2011-06-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the FimD usher bound to its cognate FimC-FimH substrate.
Nature, 474, 2011
|
|
