2KAY
 
 | | Solution structure and dynamics of S100A5 in the Ca2+ -bound states | | Descriptor: | CALCIUM ION, Protein S100-A5 | | Authors: | Bertini, I, Das Gupta, S, Hu, X, Karavelas, T, Luchinat, C, Parigi, G, Yuan, J, Structural Proteomics in Europe (SPINE), Structural Proteomics in Europe 2 (SPINE-2) | | Deposit date: | 2008-11-17 | | Release date: | 2009-06-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of S100A5 in the apo and Ca2+-bound states
J.Biol.Inorg.Chem., 14, 2009
|
|
4LBD
 
 | | LIGAND-BINDING DOMAIN OF THE HUMAN RETINOIC ACID RECEPTOR GAMMA BOUND TO THE SYNTHETIC AGONIST BMS961 | | Descriptor: | 3-FLUORO-4-[2-HYDROXY-2-(5,5,8,8-TETRAMETHYL-5,6,7,8,-TETRAHYDRO-NAPHTALEN-2-YL)-ACETYLAMINO]-BENZOIC ACID, RETINOIC ACID RECEPTOR GAMMA | | Authors: | Klaholz, B.P, Renaud, J.-P, Mitschler, A, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 1998-02-04 | | Release date: | 1999-03-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational adaptation of agonists to the human nuclear receptor RAR gamma.
Nat.Struct.Biol., 5, 1998
|
|
1QZZ
 
 | | Crystal structure of aclacinomycin-10-hydroxylase (RdmB) in complex with S-adenosyl-L-methionine (SAM) | | Descriptor: | ACETATE ION, S-ADENOSYLMETHIONINE, aclacinomycin-10-hydroxylase | | Authors: | Jansson, A, Niemi, J, Lindqvist, Y, Mantsala, P, Schneider, G, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-09-19 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Aclacinomycin-10-Hydroxylase, a S-Adenosyl-L-Methionine-dependent Methyltransferase Homolog Involved in Anthracycline Biosynthesis in Streptomyces purpurascens.
J.Mol.Biol., 334, 2003
|
|
2B1U
 
 | | Solution structure of Calmodulin-like Skin Protein C terminal domain | | Descriptor: | Calmodulin-like protein 5 | | Authors: | Babini, E, Bertini, I, Capozzi, F, Chirivino, E, Luchinat, C, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-09-16 | | Release date: | 2006-05-30 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A Structural and Dynamic Characterization of the EF-Hand Protein CLSP.
Structure, 14, 2006
|
|
2AF2
 
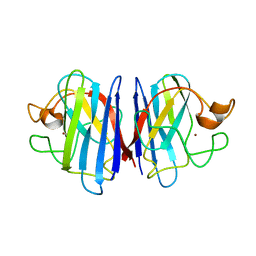 | | Solution structure of disulfide reduced and copper depleted Human Superoxide Dismutase | | Descriptor: | Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Banci, L, Bertini, I, Cantini, F, D'Amelio, N, Gaggelli, E, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-07-25 | | Release date: | 2005-11-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Human SOD1 before harboring the catalytic metal: solution structure of copper-depleted, disulfide-reduced form
J.Biol.Chem., 281, 2006
|
|
2K4W
 
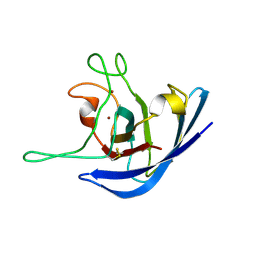 | | The Solution Structure of the Monomeric Copper, Zinc Superoxide Dismutase from Salmonella enterica | | Descriptor: | COPPER (I) ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Mori, M, Jimenez, B, Piccioli, M, Battistoni, A, Sette, M, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2008-06-20 | | Release date: | 2008-11-18 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of the Monomeric Copper, Zinc Superoxide Dismutase from Salmonella enterica: Structural Insights To Understand the Evolution toward the Dimeric Structure.
Biochemistry, 47, 2008
|
|
2LBD
 
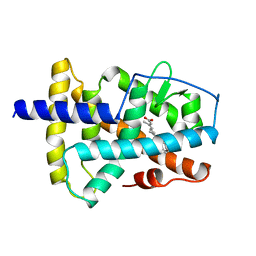 | | LIGAND-BINDING DOMAIN OF THE HUMAN RETINOIC ACID RECEPTOR GAMMA BOUND TO ALL-TRANS RETINOIC ACID | | Descriptor: | RETINOIC ACID, RETINOIC ACID RECEPTOR GAMMA | | Authors: | Renaud, J.-P, Rochel, N, Ruff, M, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 1997-08-19 | | Release date: | 1997-11-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structure of the RAR-gamma ligand-binding domain bound to all-trans retinoic acid.
Nature, 378, 1995
|
|
2RLI
 
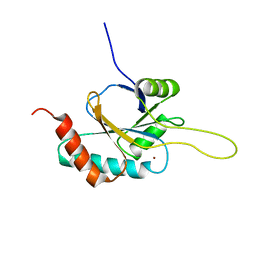 | | Solution structure of Cu(I) human Sco2 | | Descriptor: | COPPER (I) ION, SCO2 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Ciofi-baffoni, S, Gerothanassis, I.P, Leontari, I, Martinelli, M, Wang, S, Structural Proteomics in Europe (SPINE), Structural Proteomics in Europe 2 (SPINE-2) | | Deposit date: | 2007-07-11 | | Release date: | 2007-08-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A Structural Characterization of Human SCO2
Structure, 15, 2007
|
|
1BON
 
 | | THREE-DIMENSIONAL STRUCTURE OF BOMBYXIN-II, AN INSULIN-RELATED BRAIN-SECRETORY PEPTIDE OF THE SILKMOTH BOMBYX MORI: COMPARISON WITH INSULIN AND RELAXIN | | Descriptor: | BOMBYXIN-II,BOMBYXIN A-2, BOMBYXIN-II,BOMBYXIN A-6 | | Authors: | Nagata, K, Hatanaka, H, Kohda, D, Inagaki, F, Structural Proteomics in Europe (SPINE) | | Deposit date: | 1994-07-21 | | Release date: | 1995-01-26 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of bombyxin-II an insulin-like peptide of the silkmoth Bombyx mori: structural comparison with insulin and relaxin.
J.Mol.Biol., 253, 1995
|
|
3M7R
 
 | | Crystal structure of VDR H305Q mutant | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, Vitamin D3 receptor | | Authors: | Rochel, N, Hourai, S, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2010-03-17 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of hereditary vitamin D-resistant rickets--associated mutant H305Q of vitamin D nuclear receptor bound to its natural ligand
J.Steroid Biochem.Mol.Biol., 121, 2010
|
|
3LBD
 
 | | LIGAND-BINDING DOMAIN OF THE HUMAN RETINOIC ACID RECEPTOR GAMMA BOUND TO 9-CIS RETINOIC ACID | | Descriptor: | (9cis)-retinoic acid, RETINOIC ACID RECEPTOR GAMMA | | Authors: | Klaholz, B.P, Renaud, J.-P, Mitschler, A, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 1998-02-04 | | Release date: | 1999-03-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational adaptation of agonists to the human nuclear receptor RAR gamma.
Nat.Struct.Biol., 5, 1998
|
|
4FBM
 
 | | LipS and LipT, two metagenome-derived lipolytic enzymes increase the diversity of known lipase and esterase families | | Descriptor: | BROMIDE ION, LipS lipolytic enzyme | | Authors: | Chow, J, Krauss, U, Dall Antonia, Y, Fersini, F, Schmeisser, C, Schmidt, M, Menyes, I, Bornscheuer, U, Lauinger, B, Bongen, P, Pietruszka, J, Eckstein, M, Thum, O, Liese, A, Mueller-Dieckmann, J, Jaeger, K.-E, Kovavic, F, Streit, W.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2012-05-23 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Metagenome-Derived Enzymes LipS and LipT Increase the Diversity of Known Lipases.
Plos One, 7, 2012
|
|
4FBL
 
 | | LipS and LipT, two metagenome-derived lipolytic enzymes increase the diversity of known lipase and esterase families | | Descriptor: | CHLORIDE ION, LipS lipolytic enzyme, SPERMIDINE | | Authors: | Chow, J, Krauss, U, Dall Antonia, Y, Fersini, F, Schmeisser, C, Schmidt, M, Menyes, I, Bornscheuer, U, Lauinger, B, Bongen, P, Pietruszka, J, Eckstein, M, Thum, O, Liese, A, Mueller-Dieckmann, J, Jaeger, K.-E, Kovacic, F, Streit, W.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2012-05-23 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The Metagenome-Derived Enzymes LipS and LipT Increase the Diversity of Known Lipases.
Plos One, 7, 2012
|
|
2VD9
 
 | | The crystal structure of alanine racemase from Bacillus anthracis (BA0252) with bound L-Ala-P | | Descriptor: | (1S)-1-[((1E)-{3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYLENE)AMINO]ETHYLPHOSPHONIC ACID, ALANINE RACEMASE, CHLORIDE ION, ... | | Authors: | Au, K, Ren, J, Walter, T.S, Harlos, K, Nettleship, J.E, Owens, R.J, Stuart, D.I, Esnouf, R.M, Oxford Protein Production Facility (OPPF), Structural Proteomics in Europe (SPINE) | | Deposit date: | 2007-10-01 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of an Alanine Racemase from Bacillus Anthracis (Ba0252) in the Presence and Absence of (R)-1-Aminoethylphosphonic Acid (L-Ala-P).
Acta Crystallogr.,Sect.F, 64, 2008
|
|
2VD8
 
 | | The crystal structure of alanine racemase from Bacillus anthracis (BA0252) | | Descriptor: | ALANINE RACEMASE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Au, K, Ren, J, Walter, T.S, Harlos, K, Nettleship, J.E, Owens, R.J, Stuart, D.I, Esnouf, R.M, Oxford Protein Production Facility (OPPF), Structural Proteomics in Europe (SPINE) | | Deposit date: | 2007-10-01 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structures of an Alanine Racemase from Bacillus Anthracis (Ba0252) in the Presence and Absence of (R)-1-Aminoethylphosphonic Acid (L-Ala-P).
Acta Crystallogr.,Sect.F, 64, 2008
|
|
2KAX
 
 | | Solution structure and dynamics of S100A5 in the apo and Ca2+ -bound states | | Descriptor: | Protein S100-A5 | | Authors: | Bertini, I, Das Gupta, S, Hu, X, Karavelas, T, Luchinat, C, Parigi, G, Yuan, J, Structural Proteomics in Europe (SPINE), Structural Proteomics in Europe 2 (SPINE-2) | | Deposit date: | 2008-11-17 | | Release date: | 2009-06-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of S100A5 in the apo and Ca2+-bound states
J.Biol.Inorg.Chem., 14, 2009
|
|
3D24
 
 | | Crystal structure of ligand-binding domain of estrogen-related receptor alpha (ERRalpha) in complex with the peroxisome proliferators-activated receptor coactivator-1alpha box3 peptide (PGC-1alpha) | | Descriptor: | Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, Steroid hormone receptor ERR1 | | Authors: | Moras, D, Greschik, H, Flaig, R, Sato, Y, Rochel, N, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2008-05-07 | | Release date: | 2008-06-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Communication between the ERR{alpha} Homodimer Interface and the PGC-1{alpha} Binding Surface via the Helix 8-9 Loop.
J.Biol.Chem., 283, 2008
|
|
3EYB
 
 | |
3GCZ
 
 | | Yokose virus Methyltransferase in complex with AdoMet | | Descriptor: | GLYCEROL, Polyprotein, S-ADENOSYLMETHIONINE, ... | | Authors: | Bollati, M, Milani, M, Mastrangelo, E, Bolognesi, M, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2009-02-23 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a methyltransferase from a no-known-vector Flavivirus
Biochem.Biophys.Res.Commun., 382, 2009
|
|
2UVD
 
 | | The crystal structure of a 3-oxoacyl-(acyl carrier protein) reductase from Bacillus anthracis (BA3989) | | Descriptor: | 3-OXOACYL-(ACYL-CARRIER-PROTEIN) REDUCTASE | | Authors: | Zaccai, N.R, Carter, L.G, Berrow, N.S, Sainsbury, S, Nettleship, J.E, Walter, T.S, Harlos, K, Owens, R.J, Wilson, K.S, Stuart, D.I, Esnouf, R.M, Oxford Protein Production Facility (OPPF), Structural Proteomics in Europe (SPINE) | | Deposit date: | 2007-03-09 | | Release date: | 2007-04-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of a 3-Oxoacyl-(Acylcarrier Protein) Reductase (Ba3989) from Bacillus Anthracis at 2.4-A Resolution.
Proteins: Struct., Funct., Bioinf., 70, 2008
|
|
2O7P
 
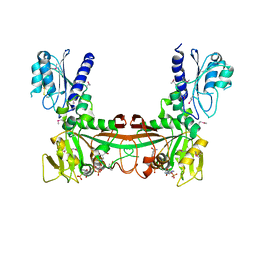 | | The crystal structure of RibD from Escherichia coli in complex with the oxidised NADP+ cofactor in the active site of the reductase domain | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Riboflavin biosynthesis protein ribD | | Authors: | Moche, M, Stenmark, P, Gurmu, D, Nordlund, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-12-11 | | Release date: | 2007-02-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of the bifunctional deaminase/reductase RibD of the riboflavin biosynthetic pathway in Escherichia coli: implications for the reductive mechanism.
J.Mol.Biol., 373, 2007
|
|
2OBC
 
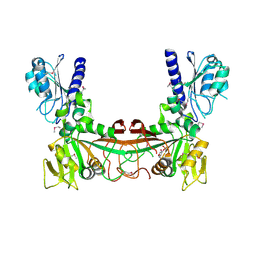 | | The crystal structure of RibD from Escherichia coli in complex with a substrate analogue, ribose 5-phosphate (beta form), bound to the active site of the reductase domain | | Descriptor: | 5-O-phosphono-beta-D-ribofuranose, Riboflavin biosynthesis protein ribD | | Authors: | Moche, M, Stenmark, P, Gurmu, D, Nordlund, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-12-18 | | Release date: | 2007-02-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of the bifunctional deaminase/reductase RibD of the riboflavin biosynthetic pathway in Escherichia coli: implications for the reductive mechanism.
J.Mol.Biol., 373, 2007
|
|
1EXA
 
 | | ENANTIOMER DISCRIMINATION ILLUSTRATED BY CRYSTAL STRUCTURES OF THE HUMAN RETINOIC ACID RECEPTOR HRARGAMMA LIGAND BINDING DOMAIN: THE COMPLEX WITH THE ACTIVE R-ENANTIOMER BMS270394. | | Descriptor: | DODECYL-ALPHA-D-MALTOSIDE, R-3-FLUORO-4-[2-HYDROXY-2-(5,5,8,8-TETRAMETHYL-5,6,7,8,-TETRAHYDRO-NAPHTALEN-2-YL)-ACETYLAMINO]-BENZOIC ACID, RETINOIC ACID RECEPTOR GAMMA-2 | | Authors: | Klaholz, B.P, Mitschler, A, Belema, M, Zusi, C, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2000-05-02 | | Release date: | 2000-06-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Enantiomer discrimination illustrated by high-resolution crystal structures of the human nuclear receptor hRARgamma.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1EXX
 
 | | ENANTIOMER DISCRIMINATION ILLUSTRATED BY CRYSTAL STRUCTURES OF THE HUMAN RETINOIC ACID RECEPTOR HRARGAMMA LIGAND BINDING DOMAIN: THE COMPLEX WITH THE INACTIVE S-ENANTIOMER BMS270395. | | Descriptor: | 3-FLUORO-4-[2-HYDROXY-2-(5,5,8,8-TETRAMETHYL-5,6,7,8,-TETRAHYDRO-NAPHTALEN-2-YL)-ACETYLAMINO]-BENZOIC ACID, DODECYL-ALPHA-D-MALTOSIDE, RETINOIC ACID RECEPTOR GAMMA-2 | | Authors: | Klaholz, B.P, Mitschler, A, Belema, M, Zusi, C, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2000-05-05 | | Release date: | 2000-06-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Enantiomer discrimination illustrated by high-resolution crystal structures of the human nuclear receptor hRARgamma.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1KV6
 
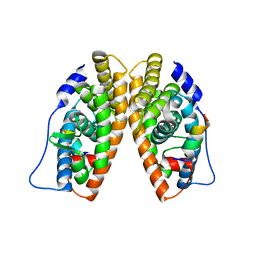 | | X-ray structure of the orphan nuclear receptor ERR3 ligand-binding domain in the constitutively active conformation | | Descriptor: | ESTROGEN-RELATED RECEPTOR GAMMA, steroid receptor coactivator 1 | | Authors: | Greschik, H, Wurtz, J.-M, Sanglier, S, Bourguet, W, van Dorsselaer, A, Moras, D, Renaud, J.-P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2002-01-25 | | Release date: | 2003-01-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Evidence for Ligand-Independent Transcriptional Activation by the Estrogen-Related Receptor 3
Mol.Cell, 9, 2002
|
|
