8IWO
 
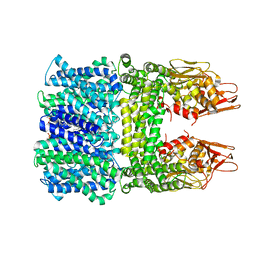 | | The rice Na+/H+ antiporter SOS1 in an auto-inhibited state | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, OsSOS1 | | Authors: | Zhang, X.Y, Tang, L.H, Zhang, C.R, Nie, J.W. | | Deposit date: | 2023-03-30 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structure and activation mechanism of the rice Salt Overly Sensitive 1 (SOS1) Na + /H + antiporter.
Nat.Plants, 9, 2023
|
|
8J2M
 
 | | The truncated rice Na+/H+ antiporter SOS1 (1-976) in a constitutively active state | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, Na+/H+ antiporter | | Authors: | Zhang, X.Y, Tang, L.H, Zhang, C.R, Nie, J.W. | | Deposit date: | 2023-04-14 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and activation mechanism of the rice Salt Overly Sensitive 1 (SOS1) Na + /H + antiporter.
Nat.Plants, 9, 2023
|
|
8YE5
 
 | |
5CF3
 
 | | Crystal structures of Bbp from Staphylococcus aureus | | Descriptor: | Bone sialoprotein-binding protein, CALCIUM ION | | Authors: | Yu, Y, Zhang, X.Y, Gu, J.K. | | Deposit date: | 2015-07-08 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.031 Å) | | Cite: | Crystal structures of Bbp from Staphylococcus aureus reveal the ligand binding mechanism with Fibrinogen alpha
Protein Cell, 6, 2015
|
|
5CFA
 
 | |
5GRM
 
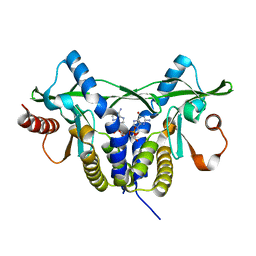 | | Crystal structure of rat STING in complex with cyclic GMP-AMP with 2'5'and 3'5'phosphodiester linkage(2'3'-cGAMP) | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Zhang, H, Han, M.J, Tao, J.L, Ye, Z.Y, Du, X.X, Deng, M.J, Zhang, X.Y, Li, L.F, Jiang, Z.F, Su, X.D. | | Deposit date: | 2016-08-11 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of rat STING in complex with cyclic GMP-AMP with 2'5'and 3'5'phosphodiester linkage(2'3'-cGAMP)
To Be Published
|
|
5GS5
 
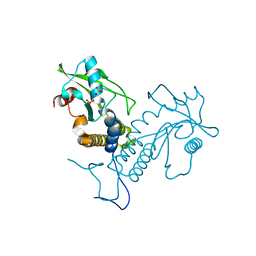 | | Crystal structure of apo rat STING | | Descriptor: | SULFATE ION, Stimulator of interferon genes protein | | Authors: | Zhang, H, Han, M.J, Tao, J.L, Ye, Z.Y, Du, X.X, Deng, M.J, Zhang, X.Y, Li, L.F, Jiang, Z.F, Su, X.D. | | Deposit date: | 2016-08-13 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of apo ratSTING
To Be Published
|
|
7Y56
 
 | |
7Y57
 
 | |
7XT1
 
 | |
7YOB
 
 | |
4XZ6
 
 | | TmoX in complex with TMAO | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Zhang, Y.Z, Li, C.Y. | | Deposit date: | 2015-02-04 | | Release date: | 2015-08-19 | | Last modified: | 2015-10-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic Insight into Trimethylamine N-Oxide Recognition by the Marine Bacterium Ruegeria pomeroyi DSS-3
J.Bacteriol., 197, 2015
|
|
4XVC
 
 | |
5XKY
 
 | | Crystal structure of DddY Se derivative | | Descriptor: | Uncharacterized protein, ZINC ION | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2017-05-10 | | Release date: | 2017-11-01 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Mechanistic Insights into Dimethylsulfoniopropionate Lyase DddY, a New Member of the Cupin Superfamily.
J. Mol. Biol., 429, 2017
|
|
6IIJ
 
 | | Cryo-EM structure of CV-A10 mature virion | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Chen, J.H, Ye, X.H, Cong, Y, Huang, Z. | | Deposit date: | 2018-10-06 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Coxsackievirus A10 atomic structure facilitating the discovery of a broad-spectrum inhibitor against human enteroviruses.
Cell Discov, 5, 2019
|
|
5GSN
 
 | | Tmm in complex with methimazole | | Descriptor: | 1-METHYL-1,3-DIHYDRO-2H-IMIDAZOLE-2-THIONE, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase, ... | | Authors: | Zhang, Y.Z, Li, C.Y. | | Deposit date: | 2016-08-16 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Structural mechanism for bacterial oxidation of oceanic trimethylamine into trimethylamine N-oxide
Mol. Microbiol., 103, 2017
|
|
5IPY
 
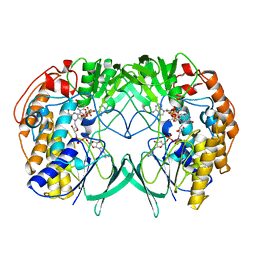 | | Crystal structure of WT RnTmm | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2016-03-10 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural mechanism for bacterial oxidation of oceanic trimethylamine into trimethylamine N-oxide
Mol. Microbiol., 103, 2017
|
|
5IQ4
 
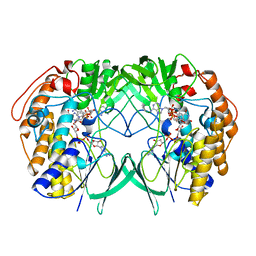 | | Crystal structure of RnTmm mutant Y207S soaking | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, Y.Z, Li, C.Y. | | Deposit date: | 2016-03-10 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural mechanism for bacterial oxidation of oceanic trimethylamine into trimethylamine N-oxide
Mol. Microbiol., 103, 2017
|
|
5IQ1
 
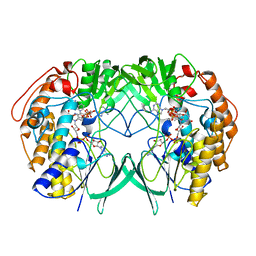 | | Crystal structure of RnTmm mutant Y207S | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2016-03-10 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural mechanism for bacterial oxidation of oceanic trimethylamine into trimethylamine N-oxide
Mol. Microbiol., 103, 2017
|
|
5GMS
 
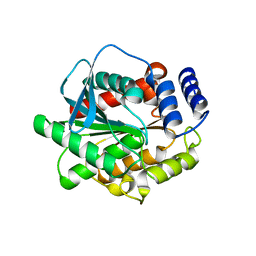 | | Crystal structure of the mutant S202W/I203F of the esterase E40 | | Descriptor: | Esterase | | Authors: | Zhang, Y.-Z, Li, P.-Y. | | Deposit date: | 2016-07-15 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Mechanistic Insights into the Improvement of the Halotolerance of a Marine Microbial Esterase by Increasing Intra- and Interdomain Hydrophobic Interactions.
Appl. Environ. Microbiol., 83, 2017
|
|
5GMR
 
 | |
4QFK
 
 | |
4QFL
 
 | |
4QFO
 
 | |
4Q05
 
 | | Crystal structure of an esterase E25 | | Descriptor: | SODIUM ION, esterase E25 | | Authors: | Li, P.Y, Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2014-03-31 | | Release date: | 2014-06-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for dimerization and catalysis of a novel esterase from the GTSAG motif subfamily of the bacterial hormone-sensitive lipase family
J.Biol.Chem., 289, 2014
|
|
