2F1N
 
 | |
2J63
 
 | | Crystal structure of AP endonuclease LMAP from Leishmania major | | Descriptor: | AP-ENDONUCLEASE | | Authors: | Vidal, A.E, Harkiolaki, M, Gallego, C, Castillo-Acosta, V.M, Ruiz-Perez, L.M, Wilson, K.S, Gonzalez-Pacanowska, D. | | Deposit date: | 2006-09-25 | | Release date: | 2007-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal Structure and DNA Repair Activities of the Ap Endonuclease from Leishmania Major.
J.Mol.Biol., 373, 2007
|
|
2ISI
 
 | |
6LPM
 
 | |
6MAL
 
 | |
5CFE
 
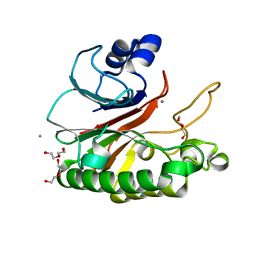 | | Bacillus subtilis AP endonuclease ExoA | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Exodeoxyribonuclease | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2015-07-08 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural comparison of AP endonucleases from the exonuclease III family reveals new amino acid residues in human AP endonuclease 1 that are involved in incision of damaged DNA.
Biochimie, 128-129, 2016
|
|
6MKM
 
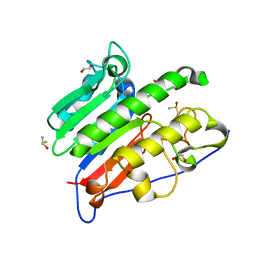 | | Crystallographic solvent mapping analysis of DMSO/Tris bound to APE1 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Georgiadis, M.M, He, H, Chen, Q. | | Deposit date: | 2018-09-25 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.673 Å) | | Cite: | Discovery of Macrocyclic Inhibitors of Apurinic/Apyrimidinic Endonuclease 1.
J. Med. Chem., 62, 2019
|
|
5CFG
 
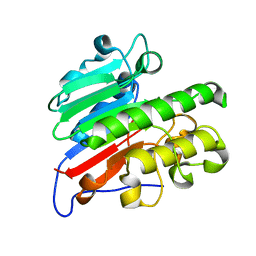 | | C2 crystal form of APE1 with Mg2+ | | Descriptor: | DNA-(apurinic or apyrimidinic site) lyase, MAGNESIUM ION | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2015-07-08 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural comparison of AP endonucleases from the exonuclease III family reveals new amino acid residues in human AP endonuclease 1 that are involved in incision of damaged DNA.
Biochimie, 128-129, 2016
|
|
6MK3
 
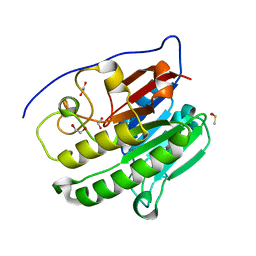 | | Crystallographic solvent mapping analysis of DMSO bound to APE1 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, DNA-(apurinic or apyrimidinic site) lyase | | Authors: | Georgiadis, M.M, He, H, Chen, Q. | | Deposit date: | 2018-09-24 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.478 Å) | | Cite: | Discovery of Macrocyclic Inhibitors of Apurinic/Apyrimidinic Endonuclease 1.
J. Med. Chem., 62, 2019
|
|
6MKO
 
 | |
6MKK
 
 | | Crystallographic solvent mapping analysis of DMSO/Mg bound to APE1 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, DNA-(apurinic or apyrimidinic site) lyase, ... | | Authors: | Georgiadis, M.M, He, H, Chen, Q. | | Deposit date: | 2018-09-25 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.442 Å) | | Cite: | Discovery of Macrocyclic Inhibitors of Apurinic/Apyrimidinic Endonuclease 1.
J. Med. Chem., 62, 2019
|
|
5DFF
 
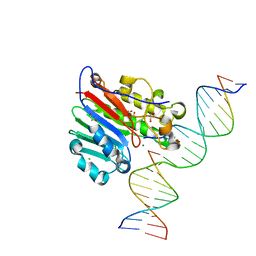 | | Human APE1 product complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Freudenthal, B.D, Wilson, S.H. | | Deposit date: | 2015-08-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Capturing snapshots of APE1 processing DNA damage.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5DFH
 
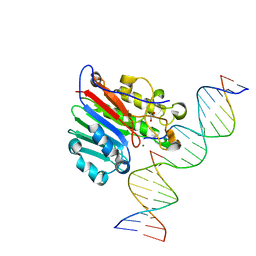 | | Human APE1 mismatch product complex | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*T)-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Freudenthal, B.D, Wilson, S.H. | | Deposit date: | 2015-08-26 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Capturing snapshots of APE1 processing DNA damage.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5DFJ
 
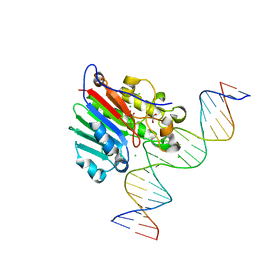 | | Human APE1 E96Q/D210N mismatch substrate complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*TP*(3DR)P*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), ... | | Authors: | Freudenthal, B.D, Wilson, S.H. | | Deposit date: | 2015-08-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Capturing snapshots of APE1 processing DNA damage.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5DG0
 
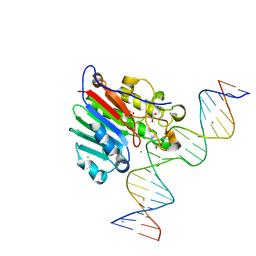 | |
5DFI
 
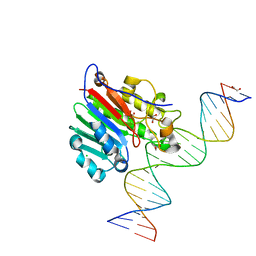 | | Human APE1 phosphorothioate substrate complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(OMC)P*(48Z)P*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), ... | | Authors: | Freudenthal, B.D, Wilson, S.H. | | Deposit date: | 2015-08-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Capturing snapshots of APE1 processing DNA damage.
Nat.Struct.Mol.Biol., 22, 2015
|
|
6NF0
 
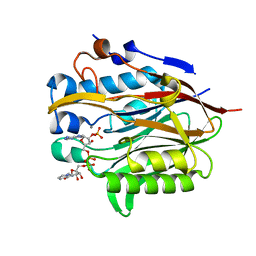 | | Nocturnin with bound NADPH substrate | | Descriptor: | CALCIUM ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Nocturnin | | Authors: | Estrella, M.A, Du, J, Korennykh, A. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The metabolites NADP+and NADPH are the targets of the circadian protein Nocturnin (Curled).
Nat Commun, 10, 2019
|
|
5DV2
 
 | |
5DV4
 
 | |
5EWT
 
 | |
6W4T
 
 | | APE1 Y269A phosphorothioate substrate complex with abasic DNA | | Descriptor: | DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*GP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*GP*CP*TP*GP*AP*TP*GP*CP*GP*TP*(48Z)P*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), DNA-(apurinic or apyrimidinic site) lyase | | Authors: | Freudenthal, B.D, Hoitsma, N.M. | | Deposit date: | 2020-03-11 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | AP-endonuclease 1 sculpts DNA through an anchoring tyrosine residue on the DNA intercalating loop.
Nucleic Acids Res., 48, 2020
|
|
6W3N
 
 | | APE1 exonuclease substrate complex D148E | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(C7R))-3'), ... | | Authors: | Freudenthal, B.D, Whitaker, A.M. | | Deposit date: | 2020-03-09 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Molecular and structural characterization of disease-associated APE1 polymorphisms.
DNA Repair (Amst.), 91-92, 2020
|
|
6W3Q
 
 | | APE1 exonuclease substrate complex L104R | | Descriptor: | CALCIUM ION, DNA-(apurinic or apyrimidinic site) lyase, GCTGATGCG(C7R), ... | | Authors: | Freudenthal, B.D, Whitaker, A.M. | | Deposit date: | 2020-03-09 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Molecular and structural characterization of disease-associated APE1 polymorphisms.
DNA Repair (Amst.), 91-92, 2020
|
|
6W2P
 
 | | APE1 endonuclease product complex L104R | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*AP*TP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Freudenthal, B.D, Whitaker, A.M. | | Deposit date: | 2020-03-06 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Molecular and structural characterization of disease-associated APE1 polymorphisms.
DNA Repair (Amst.), 91-92, 2020
|
|
6W3U
 
 | | APE1 exonuclease substrate complex R237C | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Freudenthal, B.D, Whitaker, A.M. | | Deposit date: | 2020-03-09 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular and structural characterization of disease-associated APE1 polymorphisms.
DNA Repair (Amst.), 91-92, 2020
|
|
