3ZCC
 
 | |
3ZRV
 
 | | The high resolution structure of a dimeric Hamp-Dhp fusion displays asymmetry - A291F mutant | | Descriptor: | HAMP, OSMOLARITY SENSOR PROTEIN ENVZ | | Authors: | Zeth, K, Hulko, M, Ferris, H.U, Martin, J. | | Deposit date: | 2011-06-20 | | Release date: | 2011-07-06 | | Last modified: | 2017-03-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanism of Regulation of Receptor Histidine Kinases.
Structure, 20, 2012
|
|
3ZRX
 
 | |
3ZRW
 
 | | The structure of the dimeric Hamp-Dhp fusion A291V mutant | | Descriptor: | AF1503 PROTEIN, OSMOLARITY SENSOR PROTEIN ENVZ | | Authors: | Zeth, K, Hulko, M, Ferris, H.U, Martin, J, Lupas, A.N. | | Deposit date: | 2011-06-20 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanism of Regulation of Receptor Histidine Kinases.
Structure, 20, 2012
|
|
4KP4
 
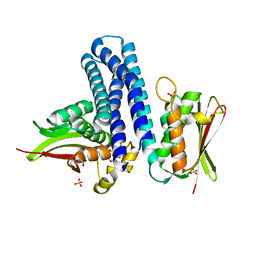 | | Deciphering cis-trans directionality and visualizing autophosphorylation in histidine kinases. | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, Osmolarity sensor protein EnvZ, ... | | Authors: | Casino, P, Miguel-Romero, L, Marina, A. | | Deposit date: | 2013-05-13 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Visualizing autophosphorylation in histidine kinases.
Nat Commun, 5, 2014
|
|
7ZP0
 
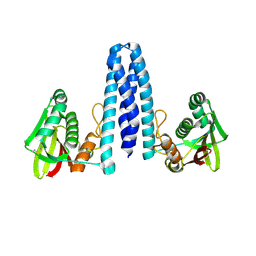 | |
7Z8N
 
 | | GacS histidine kinase from Pseudomonas aeruginosa | | Descriptor: | CALCIUM ION, Histidine kinase, R-1,2-PROPANEDIOL | | Authors: | Fadel, F, Bassim, V, Francis, V.I, Porter, S.L, Botzanowski, T, Legrand, P, Bourne, Y, Cianferani, S, Vincent, F. | | Deposit date: | 2022-03-17 | | Release date: | 2022-07-13 | | Last modified: | 2023-01-25 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Insights into the atypical autokinase activity of the Pseudomonas aeruginosa GacS histidine kinase and its interaction with RetS.
Structure, 30, 2022
|
|
8A3U
 
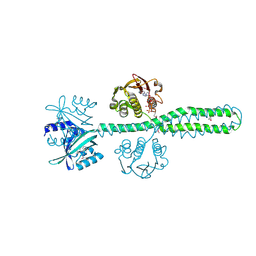 | | Crystal structure of a chimeric LOV-Histidine kinase SB2F1 (symmetrical variant, trigonal form with short c-axis) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Putative Sensory box protein,Sensor protein FixL | | Authors: | Arinkin, V, Batra-Safferling, R, Granzin, J. | | Deposit date: | 2022-06-09 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of a chimeric LOV-Histidine kinase SB2F1 (symmetrical variant, trigonal form
with short c-axis)
To Be Published
|
|
8A52
 
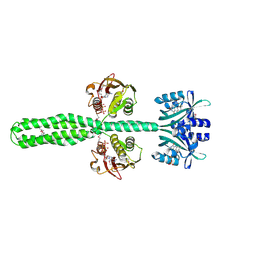 | | Crystal structure of a chimeric LOV-Histidine kinase SB2F1 (asymmetrical variant, trigonal form with long c-axis) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Putative Sensory box protein,Putative Sensory box protein,Sensor protein FixL | | Authors: | Batra-Safferling, R, Arinkin, V, Granzin, J. | | Deposit date: | 2022-06-14 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.461 Å) | | Cite: | Crystal structure of a chimeric LOV-Histidine kinase SB2F1 (asymmetrical variant, trigonal form with long c axis)
To Be Published
|
|
8A6X
 
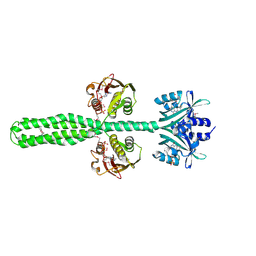 | | Crystal structure of a chimeric LOV-Histidine kinase SB2F1 (asymmetrical variant, trigonal form with long c axis) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Putative Sensory box protein,Sensor protein FixL | | Authors: | Arinkin, V, Granzin, J, Batra-Safferling, R. | | Deposit date: | 2022-06-20 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of a chimeric LOV-Histidine kinase SB2F1 (asymmetrical variant, trigonal form with long c axis)
To Be Published
|
|
8A7F
 
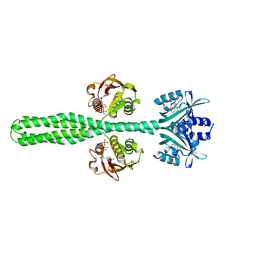 | | Crystal structure of a chimeric LOV-Histidine kinase SB2F1-I66R mutant (asymmetrical variant, trigonal form with long c axis) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Putative Sensory box protein,Sensor protein FixL | | Authors: | Batra-Safferling, R, Arinkin, V, Granzin, J. | | Deposit date: | 2022-06-21 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal structure of a chimeric LOV-Histidine kinase SB2F1-I66R mutant (asymmetrical variant, trigonal form with long c axis)
To Be Published
|
|
8A7H
 
 | | Crystal structure of a chimeric LOV-Histidine kinase SB2F1-I66R mutant (light state; asymmetrical variant, trigonal form with long c axis) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Flavin mononucleotide (semi-quinone intermediate), Putative Sensory box protein,Sensor protein FixL | | Authors: | Arinkin, V, Granzin, J, Batra-Safferling, R. | | Deposit date: | 2022-06-21 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.145 Å) | | Cite: | Crystal structure of a chimeric LOV-Histidine kinase SB2F1-I66R mutant (light state; asymmetrical variant, trigonal form with long c axis)
To Be Published
|
|
4CTI
 
 | | Escherichia coli EnvZ histidine kinase catalytic part fused to Archaeoglobus fulgidus Af1503 HAMP domain | | Descriptor: | OSMOLARITY SENSOR PROTEIN ENVZ, AF1503 | | Authors: | Ferris, H.U, Coles, M, Lupas, A.N, Hartmann, M.D. | | Deposit date: | 2014-03-13 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.847 Å) | | Cite: | Crystallographic Snapshot of the Escherichia Coli Envz Histidine Kinase in an Active Conformation.
J.Struct.Biol., 186, 2014
|
|
4BIZ
 
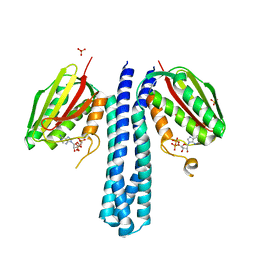 | |
4BIX
 
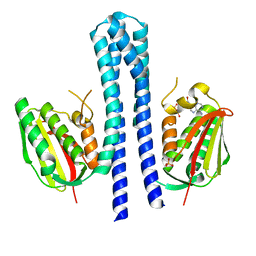 | | Crystal structure of CpxAHDC (monoclinic form 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SENSOR PROTEIN CPXA, ... | | Authors: | Mechaly, A.E, Sassoon, N, Betton, J.M, Alzari, P.M. | | Deposit date: | 2013-04-13 | | Release date: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Segmental Helical Motions and Dynamical Asymmetry Modulate Histidine Kinase Autophosphorylation.
Plos Biol., 12, 2014
|
|
4BIV
 
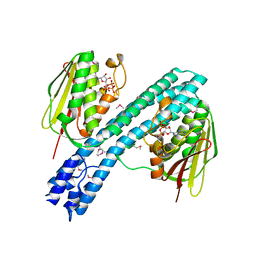 | | Crystal structure of CpxAHDC (trigonal form) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, SENSOR PROTEIN CPXA | | Authors: | Mechaly, A.E, Sassoon, N, Betton, J.M, Alzari, P.M. | | Deposit date: | 2013-04-13 | | Release date: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Segmental Helical Motions and Dynamical Asymmetry Modulate Histidine Kinase Autophosphorylation.
Plos Biol., 12, 2014
|
|
4BIW
 
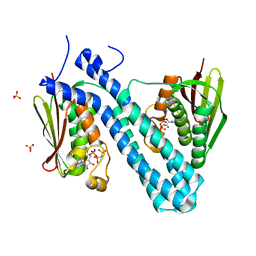 | | Crystal structure of CpxAHDC (hexagonal form) | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SENSOR PROTEIN CPXA, ... | | Authors: | Mechaly, A.E, Sassoon, N, Betton, J.M, Alzari, P.M. | | Deposit date: | 2013-04-13 | | Release date: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Segmental Helical Motions and Dynamical Asymmetry Modulate Histidine Kinase Autophosphorylation.
Plos Biol., 12, 2014
|
|
4BIU
 
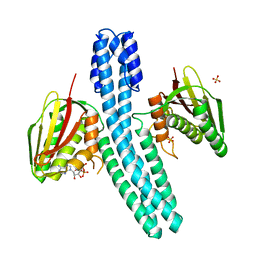 | | Crystal structure of CpxAHDC (orthorhombic form 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SENSOR PROTEIN CPXA, SULFATE ION | | Authors: | Mechaly, A.E, Sassoon, N, Betton, J.M, Alzari, P.M. | | Deposit date: | 2013-04-13 | | Release date: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Segmental Helical Motions and Dynamical Asymmetry Modulate Histidine Kinase Autophosphorylation.
Plos Biol., 12, 2014
|
|
4BIY
 
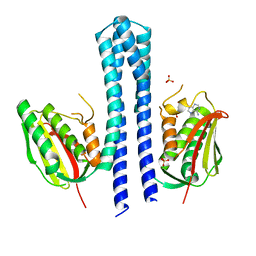 | | Crystal structure of CpxAHDC (monoclinic form 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SENSOR PROTEIN CPXA, SULFATE ION | | Authors: | Mechaly, A.E, Sassoon, N, Betton, J.M, Alzari, P.M. | | Deposit date: | 2013-04-13 | | Release date: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Segmental Helical Motions and Dynamical Asymmetry Modulate Histidine Kinase Autophosphorylation.
Plos Biol., 12, 2014
|
|
4CB0
 
 | | Crystal structure of CpxAHDC in complex with ATP (hexagonal form) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, SENSOR PROTEIN CPXA, SULFATE ION | | Authors: | Mechaly, A.E, Sassoon, N, Betton, J.M, Alzari, P.M. | | Deposit date: | 2013-10-09 | | Release date: | 2014-02-12 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Segmental Helical Motions and Dynamical Asymmetry Modulate Histidine Kinase Autophosphorylation.
Plos Biol., 12, 2014
|
|
7RZW
 
 | | CryoEM structure of Arabidopsis thaliana phytochrome B | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome B | | Authors: | Li, H, Burgie, E.S, Vierstra, R.D, Li, H. | | Deposit date: | 2021-08-27 | | Release date: | 2022-04-13 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Plant phytochrome B is an asymmetric dimer with unique signalling potential.
Nature, 604, 2022
|
|
7P8E
 
 | | Crystal structure of the Receiver domain of M. truncatula cytokinin receptor MtCRE1 | | Descriptor: | CALCIUM ION, Receiver domain of histidine kinase | | Authors: | Tran, L.H, Urbanowicz, A, Jasinski, M, Jaskolski, M, Ruszkowski, M. | | Deposit date: | 2021-07-21 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 3D Domain Swapping Dimerization of the Receiver Domain of Cytokinin Receptor CRE1 From Arabidopsis thaliana and Medicago truncatula .
Front Plant Sci, 12, 2021
|
|
3A0R
 
 | | Crystal structure of histidine kinase ThkA (TM1359) in complex with response regulator protein TrrA (TM1360) | | Descriptor: | MERCURY (II) ION, Response regulator, Sensor protein | | Authors: | Yamada, S, Sugimoto, H, Kobayashi, M, Ohno, A, Nakamura, H, Shiro, Y. | | Deposit date: | 2009-03-24 | | Release date: | 2009-10-20 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of PAS-linked histidine kinase and the response regulator complex
Structure, 17, 2009
|
|
6AZR
 
 | | Crystal structure of the T264A HK853cp-BeF3-RR468 complex | | Descriptor: | Chemotaxis regulator-transmits chemoreceptor signals to flagelllar motor components CheY, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Rose, J, Zhou, P. | | Deposit date: | 2017-09-11 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.628 Å) | | Cite: | A pH-gated conformational switch regulates the phosphatase activity of bifunctional HisKA-family histidine kinases.
Nat Commun, 8, 2017
|
|
3DGE
 
 | |
