1A2M
 
 | |
1A2J
 
 | | OXIDIZED DSBA CRYSTAL FORM II | | Descriptor: | DISULFIDE BOND FORMATION PROTEIN | | Authors: | Martin, J.L, Guddat, L.W. | | Deposit date: | 1998-01-06 | | Release date: | 1998-09-16 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of reduced and oxidized DsbA: investigation of domain motion and thiolate stabilization.
Structure, 6, 1998
|
|
1A2L
 
 | | REDUCED DSBA AT 2.7 ANGSTROMS RESOLUTION | | Descriptor: | DISULFIDE BOND FORMATION PROTEIN | | Authors: | Martin, J.L, Guddat, L.W. | | Deposit date: | 1998-01-06 | | Release date: | 1998-07-08 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of reduced and oxidized DsbA: investigation of domain motion and thiolate stabilization.
Structure, 6, 1998
|
|
1B6M
 
 | | HIV-1 PROTEASE COMPLEXED WITH MACROCYCLIC PEPTIDOMIMETIC INHIBITOR 6 | | Descriptor: | RETROPEPSIN, SULFATE ION, [1-BENZYL-3-(8-SEC-BUTYL-7,10-DIOXO-2-OXA-6,9-DIAZA-BICYCLO[11.2.2] HEPTADECA-1(16),13(17),14-TRIEN-11-YLAMINO)-2-HYDROXY-PROPYL]-CARBAMIC ACID TERT-BUTYL ESTER | | Authors: | Martin, J.L, Begun, J, Schindeler, A, Wickramasinghe, W.A, Alewood, D, Alewood, P.F, Bergman, D.A, Brinkworth, R.I, Abbenante, G, March, D.R, Reid, R.C, Fairlie, D.P. | | Deposit date: | 1999-01-17 | | Release date: | 2000-01-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular recognition of macrocyclic peptidomimetic inhibitors by HIV-1 protease.
Biochemistry, 38, 1999
|
|
1B6J
 
 | | HIV-1 PROTEASE COMPLEXED WITH MACROCYCLIC PEPTIDOMIMETIC INHIBITOR 1 | | Descriptor: | CYCLIC PEPTIDE INHIBITOR, RETROPEPSIN, SULFATE ION | | Authors: | Martin, J.L, Begun, J, Schindeler, A, Wickramasinghe, W.A, Alewood, D, Alewood, P.F, Bergman, D.A, Brinkworth, R.I, Abbenante, G, March, D.R, Reid, R.C, Fairlie, D.P. | | Deposit date: | 1999-01-17 | | Release date: | 2000-01-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular recognition of macrocyclic peptidomimetic inhibitors by HIV-1 protease.
Biochemistry, 38, 1999
|
|
1B6L
 
 | | HIV-1 PROTEASE COMPLEXED WITH MACROCYCLIC PEPTIDOMIMETIC INHIBITOR 4 | | Descriptor: | 1-[2-(8-CARBAMOYLMETHYL-6,9-DIOXO-2-OXA-7,10-DIAZA-BICYCLO[11.2.2]HEPTADECA- 1(16),13(17),14-TRIEN-11-YL)-2-HYDROXY-ETHYL]-PIPERIDINE-2-CARBOXYLIC ACID TERT-BUTYLAMIDE, RETROPEPSIN, SULFATE ION | | Authors: | Martin, J.L, Begun, J, Schindeler, A, Wickramasinghe, W.A, Alewood, D, Alewood, P.F, Bergman, D.A, Brinkworth, R.I, Abbenante, G, March, D.R, Reid, R.C, Fairlie, D.P. | | Deposit date: | 1999-01-17 | | Release date: | 2000-01-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular recognition of macrocyclic peptidomimetic inhibitors by HIV-1 protease.
Biochemistry, 38, 1999
|
|
1B6K
 
 | | HIV-1 PROTEASE COMPLEXED WITH MACROCYCLIC PEPTIDOMIMETIC INHIBITOR 5 | | Descriptor: | N-[3-(8-SEC-BUTYL-7,10-DIOXO-2-OXA-6,9-DIAZA-BICYCLO[11.2.2]HEPTADECA-1(16),13(17),14- TRIEN-11-YLAMINO)-2-HYDROXY-1-(4-HYDROXY-BENZYL)-PROPYL]-3-METHYL-2- (2-OXO-PYRROLIDIN-1-YL)-BUTYRAMIDE, RETROPEPSIN, SULFATE ION | | Authors: | Martin, J.L, Begun, J, Schindeler, A, Wickramasinghe, W.A, Alewood, D, Alewood, P.F, Bergman, D.A, Brinkworth, R.I, Abbenante, G, March, D.R, Reid, R.C, Fairlie, D.P. | | Deposit date: | 1999-01-17 | | Release date: | 2000-01-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular recognition of macrocyclic peptidomimetic inhibitors by HIV-1 protease.
Biochemistry, 38, 1999
|
|
1B6P
 
 | | HIV-1 PROTEASE COMPLEXED WITH MACROCYCLIC PEPTIDOMIMETIC INHIBITOR 7 | | Descriptor: | N-[3-(8-SEC-BUTYL-7,10-DIOXO-2-OXA-6,9-DIAZA-BICYCLO[11.2.2] HEPTADECA-1(16),13(17),14-TRIEN-11-YAMINO)-2-HYDROXY-1-(4-HYDROXY-BENZYL) -PROPYL]-3-METHYL-2-PROPIONYLAMINO-BUTYRAMIDE, RETROPEPSIN, SULFATE ION | | Authors: | Martin, J.L, Begun, J, Schindeler, A, Wickramasinghe, W.A, Alewood, D, Alewood, P.F, Bergman, D.A, Brinkworth, R.I, Abbenante, G, March, D.R, Reid, R.C, Fairlie, D.P. | | Deposit date: | 1999-01-17 | | Release date: | 2000-01-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular recognition of macrocyclic peptidomimetic inhibitors by HIV-1 protease.
Biochemistry, 38, 1999
|
|
1FVJ
 
 | |
5GPB
 
 | |
1DSB
 
 | |
1FVK
 
 | |
1DVD
 
 | | SOLUTION NMR STRUCTURE OF HUMAN STEFIN A AT PH 5.5 AND 308K, NMR, 17 STRUCTURES | | Descriptor: | STEFIN A | | Authors: | Martin, J.R, Craven, C.J, Jerala, R, Kroon-Zitko, L, Zerovnik, E, Turk, V, Waltho, J.P. | | Deposit date: | 1996-02-26 | | Release date: | 1996-08-01 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional solution structure of human stefin A.
J.Mol.Biol., 246, 1995
|
|
1DVC
 
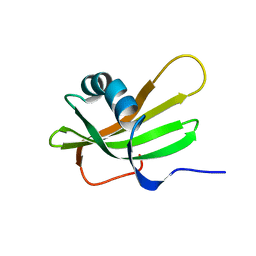 | | SOLUTION NMR STRUCTURE OF HUMAN STEFIN A AT PH 5.5 AND 308K, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | STEFIN A | | Authors: | Martin, J.R, Craven, C.J, Jerala, R, Kroon-Zitko, L, Zerovnik, E, Turk, V, Waltho, J.P. | | Deposit date: | 1996-02-26 | | Release date: | 1996-08-01 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional solution structure of human stefin A.
J.Mol.Biol., 246, 1995
|
|
1MMC
 
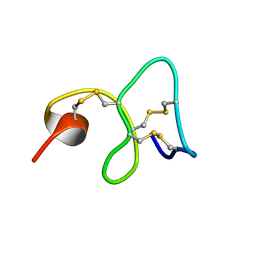 | | 1H NMR STUDY OF THE SOLUTION STRUCTURE OF AC-AMP2 | | Descriptor: | ANTIMICROBIAL PEPTIDE 2 | | Authors: | Martins, J.C, Maes, D, Loris, R, Pepermans, H.A.M, Wyns, L, Willem, R, Verheyden, P. | | Deposit date: | 1995-10-25 | | Release date: | 1996-03-08 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | H NMR study of the solution structure of Ac-AMP2, a sugar binding antimicrobial protein isolated from Amaranthus caudatus.
J.Mol.Biol., 258, 1996
|
|
1SCY
 
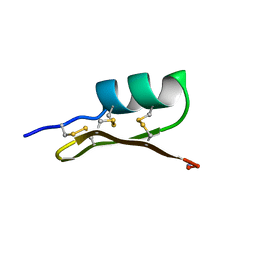 | |
1Z1H
 
 | | HIV-1 protease complexed with macrocyclic peptidomimetic inhibitor 3 | | Descriptor: | N-{(2R)-2-HYDROXY-2-[(8S,11S)-8-ISOPROPYL-6,9-DIOXO-2-OXA-7,10-DIAZABICYCLO[11.2.2]HEPTADECA-1(15),13,16-TRIEN-11-YL]ETHYL}-N-ISOPENTYLBENZENESULFONAMIDE, Pol polyprotein, SULFATE ION | | Authors: | Martin, J.L, Begun, J, Schindeler, A, Wickramasinghe, W.A, Alewood, D, Alewood, P.F, Bergman, D.A, Brinkworth, R.I, Abbenante, G, March, D.R, Reid, R.C, Fairlie, D.P. | | Deposit date: | 2005-03-04 | | Release date: | 2005-03-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular recognition of macrocyclic peptidomimetic inhibitors by HIV-1 protease
Biochemistry, 38, 1999
|
|
1Z1R
 
 | | HIV-1 protease complexed with Macrocyclic peptidomimetic inhibitor 2 | | Descriptor: | 2-[(8S,11S)-11-{(1R)-1-HYDROXY-2-[ISOPENTYL(PHENYLSULFONYL)AMINO]ETHYL}-6,9-DIOXO-2-OXA-7,10-DIAZABICYCLO[11.2.2]HEPTADECA-1(15),13,16-TRIEN-8-YL]ACETAMIDE, Pol polyprotein, SULFATE ION | | Authors: | Martin, J.L, Begun, J, Schindeler, A, Wickramasinghe, W.A, Alewood, D, Alewood, P.F, Bergman, D.A, Brinkworth, R.I, Abbenante, G, March, D.R, Reid, R.C, Fairlie, D.P. | | Deposit date: | 2005-03-06 | | Release date: | 2005-03-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular recognition of macrocyclic peptidomimetic inhibitors by HIV-1 protease
Biochemistry, 38, 1999
|
|
1HNN
 
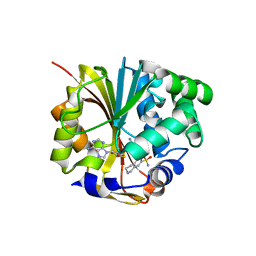 | | CRYSTAL STRUCTURE OF HUMAN PNMT COMPLEXED WITH SK&F 29661 AND ADOHCY(SAH) | | Descriptor: | 1,2,3,4-TETRAHYDRO-ISOQUINOLINE-7-SULFONIC ACID AMIDE, PHENYLETHANOLAMINE N-METHYLTRANSFERASE, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Martin, J.L, Begun, J, McLeish, M.J, Caine, J.M, Grunewald, G.L. | | Deposit date: | 2000-12-07 | | Release date: | 2001-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Getting the adrenaline going: crystal structure of the adrenaline-synthesizing enzyme PNMT.
Structure, 9, 2001
|
|
1HTX
 
 | | SOLUTION STRUCTURE OF THE MAIN ALPHA-AMYLASE INHIBITOR FROM AMARANTH SEEDS | | Descriptor: | ALPHA-AMYLASE INHIBITOR AAI | | Authors: | Martins, J.C, Enassar, M, Willem, R, Wieruzeski, J.M, Lippens, G, Wodak, S.J. | | Deposit date: | 2001-01-02 | | Release date: | 2001-07-18 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the main alpha-amylase inhibitor from amaranth seeds.
Eur.J.Biochem., 268, 2001
|
|
3E90
 
 | |
1CPI
 
 | |
3GPB
 
 | |
6R5Z
 
 | |
6R5Y
 
 | |
