4JSP
 
 | | structure of mTORDeltaN-mLST8-ATPgammaS-Mg complex | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase mTOR, ... | | Authors: | Pavletich, N.P, Yang, H. | | Deposit date: | 2013-03-22 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | mTOR kinase structure, mechanism and regulation.
Nature, 497, 2013
|
|
5SWR
 
 | | Crystal Structure of PI3Kalpha in complex with fragments 20 and 26 | | Descriptor: | 2-HYDROXYBENZOIC ACID, 6-hydroxy-3,4-dihydronaphthalen-1(2H)-one, CHLORIDE ION, ... | | Authors: | Gabelli, S.B, Vogelstein, B, Miller, M.S, Amzel, L.M. | | Deposit date: | 2016-08-08 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Identification of allosteric binding sites for PI3K alpha oncogenic mutant specific inhibitor design.
Bioorg. Med. Chem., 25, 2017
|
|
4WAE
 
 | | Phosphatidylinositol 4-kinase III beta crystallized with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Phosphatidylinositol 4-kinase beta,Phosphatidylinositol 4-kinase beta | | Authors: | Chalupska, D, Boura, E. | | Deposit date: | 2014-08-29 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.318 Å) | | Cite: | Highly Selective Phosphatidylinositol 4-Kinase III beta Inhibitors and Structural Insight into Their Mode of Action.
J.Med.Chem., 58, 2015
|
|
8SOA
 
 | | Phosphoinositide phosphate 3 kinase gamma bound with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Phosphoinositide 3-kinase regulatory subunit 5, phosphatidylinositol-4,5-bisphosphate 3-kinase | | Authors: | Chen, C.-L, Tesmer, J.J.G. | | Deposit date: | 2023-04-28 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Molecular basis for G beta gamma-mediated activation of phosphoinositide 3-kinase gamma.
Nat.Struct.Mol.Biol., 2024
|
|
7TYR
 
 | |
7OTM
 
 | | Cryo-EM structure of DNA-PKcs in complex with NU7441 | | Descriptor: | 8-(dibenzo[b,d]thiophen-4-yl)-2-(morpholin-4-yl)-4H-chromen-4-one, DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Liang, S, Thomas, S.E, Blundell, T.L. | | Deposit date: | 2021-06-10 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structural insights into inhibitor regulation of the DNA repair protein DNA-PKcs.
Nature, 601, 2022
|
|
7Z88
 
 | | DNA-PK in the intermediate state | | Descriptor: | (~{S})-[2-chloranyl-4-fluoranyl-5-(7-morpholin-4-ylquinazolin-4-yl)phenyl]-(6-methoxypyridazin-3-yl)methanol, DNA (26-MER), DNA-dependent protein kinase catalytic subunit, ... | | Authors: | Liang, S, Blundell, T.L. | | Deposit date: | 2022-03-16 | | Release date: | 2023-01-18 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Human DNA-dependent protein kinase activation mechanism.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6NCT
 
 | | Structure of p110alpha/niSH2 - vector data collection | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, SULFATE ION, ... | | Authors: | Miller, M.S, Maheshwari, S, Amzel, L.M, Gabelli, S.B. | | Deposit date: | 2018-12-12 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Getting the Most Out of Your Crystals: Data Collection at the New High-Flux, Microfocus MX Beamlines at NSLS-II.
Molecules, 24, 2019
|
|
5SXA
 
 | | Crystal Structure of PI3Kalpha in complex with fragment 10 | | Descriptor: | 2-(trifluoromethyl)-1H-benzimidazol-5-amine, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Gabelli, S.B, Vogelstein, B, Miller, M.S, Amzel, L.M. | | Deposit date: | 2016-08-09 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Identification of allosteric binding sites for PI3K alpha oncogenic mutant specific inhibitor design.
Bioorg. Med. Chem., 25, 2017
|
|
8V8J
 
 | | PI3Ka H1047R co-crystal structure with inhibitors in two cryptic pockets (compounds 4 and 5). | | Descriptor: | (2S)-N~1~-[(4P)-2-tert-butyl-4'-methyl[4,5'-bi-1,3-thiazol]-2'-yl]pyrrolidine-1,2-dicarboxamide, 2-({(1R)-1-[2-(4,4-dimethylpiperidin-1-yl)-3,6-dimethyl-4-oxo-4H-1-benzopyran-8-yl]ethyl}amino)benzoic acid, N-{(3S)-3-(2-methylphenyl)-6-[(oxetan-3-yl)amino]-1-oxo-2,3-dihydro-1H-isoindol-4-yl}-1-benzothiophene-3-carboxamide, ... | | Authors: | Gunn, R.J, Lawson, J.D. | | Deposit date: | 2023-12-05 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Discovery of Pyridopyrimidinones that Selectively Inhibit the H1047R PI3K alpha Mutant Protein.
J.Med.Chem., 67, 2024
|
|
5FBL
 
 | | PI4KB in complex with Rab11 and the MI356 Inhibitor | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Phosphatidylinositol 4-kinase beta,Phosphatidylinositol 4-kinase beta, Ras-related protein Rab-11A, ... | | Authors: | Chalupska, D, Mejdrova, I, Nencka, R, Boura, E. | | Deposit date: | 2015-12-14 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.372 Å) | | Cite: | PI4KB in complex with Rab11 and the MI356 Inhibitor
To Be Published
|
|
8SOD
 
 | |
7PW5
 
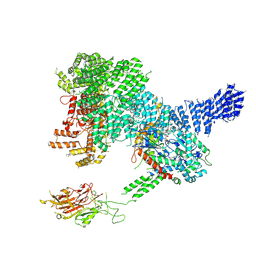 | |
5SXI
 
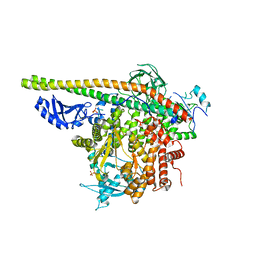 | | Crystal Structure of PI3Kalpha in complex with fragment 13 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, trans-cyclohexane-1,4-diol | | Authors: | Gabelli, S.B, Vogelstein, B, Miller, M.S, Amzel, L.M. | | Deposit date: | 2016-08-09 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Identification of allosteric binding sites for PI3K alpha oncogenic mutant specific inhibitor design.
Bioorg. Med. Chem., 25, 2017
|
|
4G11
 
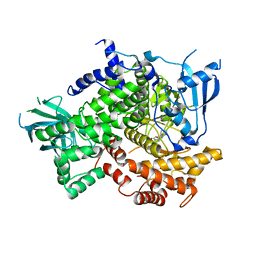 | |
2X6I
 
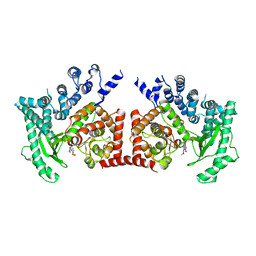 | | THE CRYSTAL STRUCTURE OF THE DROSOPHILA CLASS III PI3-KINASE VPS34 IN COMPLEX WITH PIK-90 | | Descriptor: | N-(2,3-DIHYDRO-7,8-DIMETHOXYIMIDAZO[1,2-C] QUINAZOLIN-5-YL)NICOTINAMIDE, PHOSPHOTIDYLINOSITOL 3 KINASE 59F | | Authors: | Miller, S, Tavshanjian, B, Oleksy, A, Perisic, O, Houseman, B.T, Shokat, K.M, Williams, R.L. | | Deposit date: | 2010-02-17 | | Release date: | 2010-04-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Shaping Development of Autophagy Inhibitors with the Structure of the Lipid Kinase Vps34.
Science, 327, 2010
|
|
8DD8
 
 | | PI 3-kinase alpha with nanobody 3-142, crosslinked with DSG | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7OTP
 
 | | DNA-PKcs in complex with ATPgammaS-Mg | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Liang, S, Thomas, S.E, Blundell, T.L. | | Deposit date: | 2021-06-10 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into inhibitor regulation of the DNA repair protein DNA-PKcs.
Nature, 601, 2022
|
|
6SKZ
 
 | | Structure of the closed conformation of CtTel1 | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase Tel1 | | Authors: | Jansma, M, Eustermann, S.E, Kostrewa, D, Lammens, K, Hopfner, K.P. | | Deposit date: | 2019-08-16 | | Release date: | 2019-10-30 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Near-Complete Structure and Model of Tel1ATM from Chaetomium thermophilum Reveals a Robust Autoinhibited ATP State.
Structure, 28, 2020
|
|
6T9J
 
 | | SAGA Tra1 module | | Descriptor: | Transcription factor SPT20, Transcription initiation factor TFIID subunit 12, Transcription-associated protein 1 | | Authors: | Wang, H, Cheung, A, Cramer, P. | | Deposit date: | 2019-10-28 | | Release date: | 2020-01-29 | | Last modified: | 2020-02-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the transcription coactivator SAGA.
Nature, 577, 2020
|
|
7ZVW
 
 | | NuA4 Histone Acetyltransferase Complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Chromatin modification-related protein EAF1, ... | | Authors: | Schultz, P, Ben-Shem, A, Frechard, A. | | Deposit date: | 2022-05-17 | | Release date: | 2023-05-31 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The structure of the NuA4-Tip60 complex reveals the mechanism and importance of long-range chromatin modification.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4WAG
 
 | | Phosphatidylinositol 4-kinase III beta crystallized with MI103 inhibitor | | Descriptor: | 6-chloro-3-(3,4-dimethoxyphenyl)-2-methylimidazo[1,2-b]pyridazin-8-amine, Phosphatidylinositol 4-kinase beta,Phosphatidylinositol 4-kinase beta | | Authors: | Chalupska, D, Boura, E. | | Deposit date: | 2014-08-29 | | Release date: | 2015-05-20 | | Last modified: | 2017-09-06 | | Method: | X-RAY DIFFRACTION (3.407 Å) | | Cite: | Highly Selective Phosphatidylinositol 4-Kinase III beta Inhibitors and Structural Insight into Their Mode of Action.
J.Med.Chem., 58, 2015
|
|
5SWP
 
 | | Crystal Structure of PI3Kalpha in complex with fragments 6 and 24 | | Descriptor: | 2-methylcyclohexane-1,3-dione, CHLORIDE ION, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Gabelli, S.B, Vogelstein, B, Miller, M.S, Amzel, L.M. | | Deposit date: | 2016-08-08 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Identification of allosteric binding sites for PI3K alpha oncogenic mutant specific inhibitor design.
Bioorg. Med. Chem., 25, 2017
|
|
7PE7
 
 | |
5SXJ
 
 | | Crystal Structure of PI3Kalpha in complex with fragment 29 | | Descriptor: | BENZHYDROXAMIC ACID, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Gabelli, S.B, Vogelstein, B, Miller, M.S, Amzel, L.M. | | Deposit date: | 2016-08-09 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Identification of allosteric binding sites for PI3K alpha oncogenic mutant specific inhibitor design.
Bioorg. Med. Chem., 25, 2017
|
|
