5KYD
 
 | |
5KYC
 
 | |
5KYB
 
 | |
5K1C
 
 | | Crystal structure of the UAF1/WDR20/USP12 complex | | Descriptor: | PHOSPHATE ION, TRIS(HYDROXYETHYL)AMINOMETHANE, Ubiquitin carboxyl-terminal hydrolase 12, ... | | Authors: | Li, H, D'Andrea, A.D, Zheng, N. | | Deposit date: | 2016-05-18 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Allosteric Activation of Ubiquitin-Specific Proteases by beta-Propeller Proteins UAF1 and WDR20.
Mol.Cell, 63, 2016
|
|
5K1B
 
 | |
5K1A
 
 | |
5K16
 
 | |
5JTV
 
 | | USP7CD-UBL45 in complex with Ubiquitin | | Descriptor: | Polyubiquitin-B, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Murray, J.M, Rouge, L. | | Deposit date: | 2016-05-09 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.312 Å) | | Cite: | Molecular Understanding of USP7 Substrate Recognition and C-Terminal Activation.
Structure, 24, 2016
|
|
5JTJ
 
 | | USP7CD-CTP in complex with Ubiquitin | | Descriptor: | CALCIUM ION, Polyubiquitin-B, Ubiquitin carboxyl-terminal hydrolase 7,Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Murray, J.M, Rouge, L. | | Deposit date: | 2016-05-09 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.321 Å) | | Cite: | Molecular Understanding of USP7 Substrate Recognition and C-Terminal Activation.
Structure, 24, 2016
|
|
5J7T
 
 | |
5GJQ
 
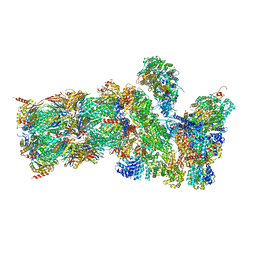 | | Structure of the human 26S proteasome bound to USP14-UbAl | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Huang, X.L, Luan, B, Wu, J.P, Shi, Y.G. | | Deposit date: | 2016-07-01 | | Release date: | 2016-08-17 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | An atomic structure of the human 26S proteasome.
Nat. Struct. Mol. Biol., 23, 2016
|
|
5FWI
 
 | |
5CVO
 
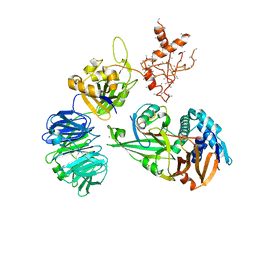 | | WDR48:USP46~ubiquitin ternary complex | | Descriptor: | Polyubiquitin-B, Ubiquitin carboxyl-terminal hydrolase 46, WD repeat-containing protein 48, ... | | Authors: | Harris, S.F, Yin, J. | | Deposit date: | 2015-07-27 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.885 Å) | | Cite: | Structural Insights into WD-Repeat 48 Activation of Ubiquitin-Specific Protease 46.
Structure, 23, 2015
|
|
5CVN
 
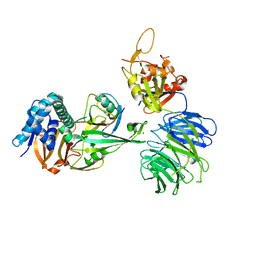 | | WDR48 (2-580):USP46~ubiquitin ternary complex | | Descriptor: | Polyubiquitin-B, Ubiquitin carboxyl-terminal hydrolase 46, WD repeat-containing protein 48, ... | | Authors: | Harris, S.F, Yin, J. | | Deposit date: | 2015-07-27 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Structural Insights into WD-Repeat 48 Activation of Ubiquitin-Specific Protease 46.
Structure, 23, 2015
|
|
5CVM
 
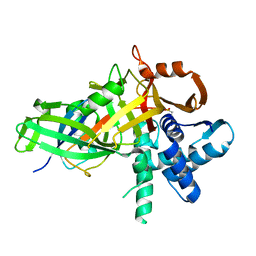 | | USP46~ubiquitin BEA covalent complex | | Descriptor: | Polyubiquitin-B, Ubiquitin carboxyl-terminal hydrolase 46, ZINC ION | | Authors: | Harris, S.F, Yin, J. | | Deposit date: | 2015-07-27 | | Release date: | 2015-10-07 | | Last modified: | 2015-11-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into WD-Repeat 48 Activation of Ubiquitin-Specific Protease 46.
Structure, 23, 2015
|
|
5CHV
 
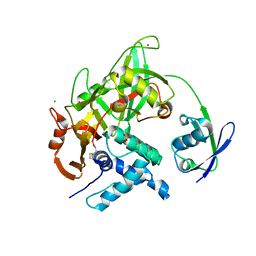 | | Crystal structure of USP18-ISG15 complex | | Descriptor: | CHLORIDE ION, SULFATE ION, Ubiquitin-like protein ISG15, ... | | Authors: | Fritz, G, Basters, A. | | Deposit date: | 2015-07-10 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Structural basis of the specificity of USP18 toward ISG15.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5CHT
 
 | | Crystal structure of USP18 | | Descriptor: | Ubl carboxyl-terminal hydrolase 18, ZINC ION | | Authors: | Fritz, G, Basters, A. | | Deposit date: | 2015-07-10 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of the specificity of USP18 toward ISG15.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5A5B
 
 | | Structure of the 26S proteasome-Ubp6 complex | | Descriptor: | 26S PROTEASE REGULATORY SUBUNIT 4 HOMOLOG, 26S PROTEASE REGULATORY SUBUNIT 6A, 26S PROTEASE REGULATORY SUBUNIT 6B HOMOLOG, ... | | Authors: | Aufderheide, A, Beck, F, Stengel, F, Hartwig, M, Schweitzer, A, Pfeifer, G, Goldberg, A.L, Sakata, E, Baumeister, W, Foerster, F. | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-22 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Structural Characterization of the Interaction of Ubp6 with the 26S Proteasome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4ZUX
 
 | |
4WA6
 
 | |
4W4U
 
 | |
4MSX
 
 | | Crystal structure of an essential yeast splicing factor | | Descriptor: | ACETATE ION, Pre-mRNA-splicing factor SAD1, ZINC ION | | Authors: | Hadjivassiliou, H, Guthrie, C, Rosenberg, O.S. | | Deposit date: | 2013-09-18 | | Release date: | 2014-04-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The crystal structure of S. cerevisiae Sad1, a catalytically inactive deubiquitinase that is broadly required for pre-mRNA splicing.
Rna, 20, 2014
|
|
4M5X
 
 | | Crystal structure of the USP7/HAUSP catalytic domain | | Descriptor: | BROMIDE ION, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Mesecar, A.D, Molland, K.L, Zhou, Q. | | Deposit date: | 2013-08-08 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | A 2.2 angstrom resolution structure of the USP7 catalytic domain in a new space group elaborates upon structural rearrangements resulting from ubiquitin binding.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
4M5W
 
 | | Crystal structure of the USP7/HAUSP catalytic domain | | Descriptor: | BROMIDE ION, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Molland, K.L, Mesecar, A.D, Zhou, Q. | | Deposit date: | 2013-08-08 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | A 2.2 angstrom resolution structure of the USP7 catalytic domain in a new space group elaborates upon structural rearrangements resulting from ubiquitin binding.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
4FK5
 
 | | Structure of the SAGA Ubp8(S144N)/Sgf11/Sus1/Sgf73 DUB module | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein SUS1, ... | | Authors: | Samara, N.L, Ringel, A.E, Wolberger, C. | | Deposit date: | 2012-06-12 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.032 Å) | | Cite: | A Role for Intersubunit Interactions in Maintaining SAGA Deubiquitinating Module Structure and Activity.
Structure, 20, 2012
|
|
