4ZUX
 
 | |
6AQR
 
 | | SAGA DUB module Ubp8(C146A)/Sgf11/Sus1/Sgf73 bound to monoubiquitin | | Descriptor: | Polyubiquitin-C, SAGA-associated factor 11, SAGA-associated factor 73, ... | | Authors: | Morrow, M.E, Morgan, M.T, Wolberger, C. | | Deposit date: | 2017-08-21 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Active site alanine mutations convert deubiquitinases into high-affinity ubiquitin-binding proteins.
EMBO Rep., 19, 2018
|
|
2KBM
 
 | | Ca-S100A1 interacting with TRTK12 | | Descriptor: | CALCIUM ION, F-actin-capping protein subunit alpha-2, Protein S100-A1 | | Authors: | Wright, N.T, Varney, K.M, Cannon, B.R, Morgan, M, Weber, D.J. | | Deposit date: | 2008-12-02 | | Release date: | 2009-02-10 | | Last modified: | 2018-08-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Ca-S100A1-TRTK12
To be Published
|
|
2CJM
 
 | | Mechanism of CDK inhibition by active site phosphorylation: CDK2 Y15p T160p in complex with cyclin A structure | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, ... | | Authors: | Welburn, J.P.I, Tucker, J, Johnson, T, Lindert, L, Morgan, M, Willis, A, Noble, M.E.M, Endicott, J.A. | | Deposit date: | 2006-04-05 | | Release date: | 2006-04-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | How Tyrosine 15 Phosphorylation Inhibits the Activity of Cyclin-Dependent Kinase 2-Cyclin A.
J.Biol.Chem., 282, 2007
|
|
5J4K
 
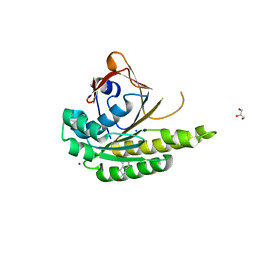 | | Structure of humanised RadA-mutant humRadA22F in complex with 1-Indane-6-carboxylic acid | | Descriptor: | 2,3-dihydro-1H-indene-2-carboxylic acid, CALCIUM ION, DNA repair and recombination protein RadA, ... | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-01 | | Release date: | 2016-10-26 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (1.346 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5JEC
 
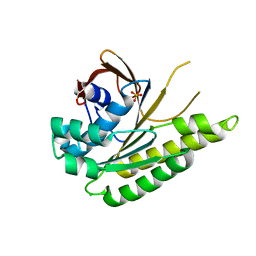 | | Apo-structure of humanised RadA-mutant humRadA33F | | Descriptor: | CHLORIDE ION, DNA repair and recombination protein RadA, SULFATE ION | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-18 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5J4H
 
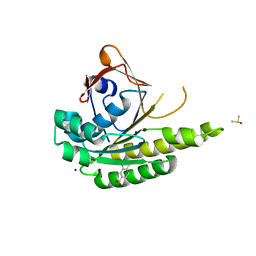 | | Structure of humanised RadA-mutant humRadA22F in complex with indole-6-carboxylic acid | | Descriptor: | 1H-indole-6-carboxylic acid, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-01 | | Release date: | 2016-10-19 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (1.374 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5JED
 
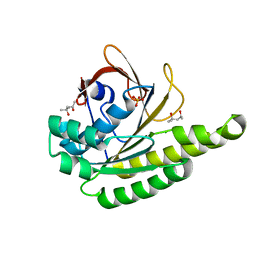 | | Apo-structure of humanised RadA-mutant humRadA28 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-18 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.332 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5J4L
 
 | | Apo-structure of humanised RadA-mutant humRadA22F | | Descriptor: | CHLORIDE ION, DNA repair and recombination protein RadA | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-01 | | Release date: | 2016-10-19 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5JEE
 
 | | Apo-structure of humanised RadA-mutant humRadA26F | | Descriptor: | CALCIUM ION, DNA repair and recombination protein RadA | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-18 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5JFG
 
 | | Structure of humanised RadA-mutant humRadA22F in complex with peptide FHTA | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, DNA repair and recombination protein RadA, ... | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-19 | | Release date: | 2016-10-26 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5L8V
 
 | | Apo-structure of humanised RadA-mutant humRadA4 | | Descriptor: | DNA repair and recombination protein RadA, PHOSPHATE ION | | Authors: | Marsh, M, Fischer, G, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-06-08 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5LB4
 
 | | Apo-structure of humanised RadA-mutant humRadA14 | | Descriptor: | DNA repair and recombination protein RadA | | Authors: | Marsh, M, Fischer, G, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-06-15 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
6SHG
 
 | | Diffraction data for RoAb13 crystal co-crystallised with PIYDIN and its RoAb13 structure | | Descriptor: | Antibody RoAb13 Heavy Chain, Antibody RoAb13 Light Chain | | Authors: | Saridakis, E, Helliwell, J.R, Govada, L, Chain, B, Morgan, M, Kassen, S.C, Chayen, N.E. | | Deposit date: | 2019-08-06 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.35050821 Å) | | Cite: | X-ray crystallography based model of the antibody RoAb13 bound to peptidomimetics of the HIV receptor chemokine receptor CCR5
To Be Published
|
|
5KDD
 
 | | Apo-structure of humanised RadA-mutant humRadA22 | | Descriptor: | DNA repair and recombination protein RadA, SULFATE ION | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-06-08 | | Release date: | 2016-10-19 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5LBI
 
 | | Apo-structure of humanised RadA-mutant humRadA3 | | Descriptor: | CALCIUM ION, DNA repair and recombination protein RadA | | Authors: | Marsh, M, Fischer, G, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-06-16 | | Release date: | 2016-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5LB2
 
 | | Apo-structure of humanised RadA-mutant humRadA2 | | Descriptor: | DNA repair and recombination protein RadA | | Authors: | Marsh, M, Fischer, G, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-06-15 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
6FW7
 
 | | Crystal structure of L-tryptophan oxidase VioA from Chromobacterium violaceum in complex with 4-Fluoro-L-Tryptophan | | Descriptor: | 4-FLUOROTRYPTOPHANE, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent L-tryptophan oxidase VioA, ... | | Authors: | Lai, H.E, Morgan, M, Moore, S, Freemont, P. | | Deposit date: | 2018-03-05 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A GenoChemetic strategy for derivatization of the violacein natural product scaffold
Biorxiv, 2019
|
|
6FW9
 
 | | Crystal structure of L-tryptophan oxidase VioA from Chromobacterium violaceum in complex with 6-Fluoro-L-Tryptophan | | Descriptor: | 6-FLUORO-L-TRYPTOPHAN, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent L-tryptophan oxidase VioA, ... | | Authors: | Lai, H.E, Morgan, M, Moore, S, Freemont, P. | | Deposit date: | 2018-03-05 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.739 Å) | | Cite: | A GenoChemetic strategy for derivatization of the violacein natural product scaffold
Biorxiv, 2019
|
|
6FWA
 
 | | Crystal structure of L-tryptophan oxidase VioA from Chromobacterium violaceum in complex with 7-Methyl-L-Tryptophan | | Descriptor: | 7-Methyl-L-Tryptophan, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent L-tryptophan oxidase VioA, ... | | Authors: | Lai, H.E, Morgan, M, Moore, S, Freemont, P. | | Deposit date: | 2018-03-05 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.846 Å) | | Cite: | A GenoChemetic strategy for derivatization of the violacein natural product scaffold
Biorxiv, 2019
|
|
6FW8
 
 | | Crystal structure of L-tryptophan oxidase VioA from Chromobacterium violaceum in complex with 5-Methyl-L-Tryptophan | | Descriptor: | 5-methyl-L-tryptophan, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent L-tryptophan oxidase VioA, ... | | Authors: | Lai, H.E, Morgan, M, Moore, S, Freemont, P. | | Deposit date: | 2018-03-05 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A GenoChemetic strategy for derivatization of the violacein natural product scaffold
Biorxiv, 2019
|
|
6G2P
 
 | | Crystal structure of L-tryptophan oxidase VioA from Chromobacterium violaceum in complex with L-tryptophan | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent L-tryptophan oxidase VioA, ... | | Authors: | Lai, H.E, Morgan, M, Moore, S, Freemont, P. | | Deposit date: | 2018-03-23 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A semi-synthetic strategy for derivatization of the violacein natural product scaffold
Biorxiv, 2018
|
|
6ESD
 
 | | Crystal structure of L-tryptophan oxidase VioA from Chromobacterium violaceum | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent L-tryptophan oxidase VioA | | Authors: | Lai, H.E, Morgan, M, Moore, S, Freemont, P. | | Deposit date: | 2017-10-20 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A semi-synthetic strategy for derivatization of the violacein natural product scaffold
Biorxiv, 2017
|
|
6GV0
 
 | | Insulin glulisine | | Descriptor: | FORMIC ACID, Insulin, ZINC ION | | Authors: | Chayen, N.E, Helliwell, J.R, Solomon-Gamsu, H.V, Govada, L, Morgan, M, Gillis, R.B, Adams, G. | | Deposit date: | 2018-06-20 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Analysis of insulin glulisine at the molecular level by X-ray crystallography and biophysical techniques.
Sci Rep, 11, 2021
|
|
