8SDL
 
 | | Crystal structure of PDC-3 beta-lactamase | | Descriptor: | Beta-lactamase, IMIDAZOLE, ISOPROPYL ALCOHOL | | Authors: | Kumar, V, van den Akker, F. | | Deposit date: | 2023-04-07 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Natural protein engineering in the Omega-loop: the role of Y221 in ceftazidime and ceftolozane resistance in Pseudomonas -derived cephalosporinase.
Antimicrob.Agents Chemother., 67, 2023
|
|
3WS2
 
 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (Condition-1C) | | Descriptor: | Beta-lactamase, CALCIUM ION, CESIUM ION | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3W8K
 
 | | Crystal structure of class C beta-lactamase Mox-1 | | Descriptor: | ACETATE ION, Beta-lactamase, ZINC ION | | Authors: | Shimizu-ibuka, A, Oguri, T, Furuyama, T, Ishii, Y. | | Deposit date: | 2013-03-15 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Mox-1, a unique plasmid-mediated class C beta-lactamase with hydrolytic activity towards moxalactam
Antimicrob.Agents Chemother., 58, 2014
|
|
3WS1
 
 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (Condition-1B) | | Descriptor: | Beta-lactamase, CALCIUM ION, CESIUM ION | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8RLJ
 
 | | Structure of the apo form of PIB-1 in an Orthorombic space group | | Descriptor: | Class C beta-lactamase-related serine hydrolase | | Authors: | Medrano, F.J, Romero, A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A new type of Class C beta-lactamases defined by PIB-1. A metal-dependent carbapenem-hydrolyzing beta-lactamase, from Pseudomonas aeruginosa: Structural and functional analysis.
Int.J.Biol.Macromol., 277, 2024
|
|
8RLL
 
 | | Structure of the apo form of PIB-1 in an Orthorombic space group | | Descriptor: | Class C beta-lactamase-related serine hydrolase | | Authors: | Medrano, F.J, Romero, A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | A new type of Class C beta-lactamases defined by PIB-1. A metal-dependent carbapenem-hydrolyzing beta-lactamase, from Pseudomonas aeruginosa: Structural and functional analysis.
Int.J.Biol.Macromol., 277, 2024
|
|
8RLK
 
 | | Structure of the apo form of PIB-1 in an Orthorombic space group | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, Class C beta-lactamase-related serine hydrolase, MAGNESIUM ION, ... | | Authors: | Medrano, F.J, Romero, A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A new type of Class C beta-lactamases defined by PIB-1. A metal-dependent carbapenem-hydrolyzing beta-lactamase, from Pseudomonas aeruginosa: Structural and functional analysis.
Int.J.Biol.Macromol., 277, 2024
|
|
3WS5
 
 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (Condition-2B) | | Descriptor: | Beta-lactamase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-28 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8TTP
 
 | | Crystal structure of class C beta-lactamase from Escherichia coli in complex with avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1,2-ETHANEDIOL, Beta-lactamase, ... | | Authors: | Chang, C, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-08-14 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal structure of class C beta-lactamase from Escherichia coli in complex with avibactam
to be published
|
|
4BLM
 
 | |
3ZYT
 
 | |
3WS0
 
 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (Condition-1A) | | Descriptor: | Beta-lactamase, CALCIUM ION, CESIUM ION, ... | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WWX
 
 | | Crystal structure of D-stereospecific amidohydrolase from Streptomyces sp. 82F2 | | Descriptor: | OCTANE 1,8-DIAMINE, S12 family peptidase | | Authors: | Arima, J, Nagano, S, Hino, T, Shimone, K, Isoda, Y, Mori, N. | | Deposit date: | 2014-07-03 | | Release date: | 2015-07-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structure of D-stereospecific amidohydrolase from Streptomyces sp. 82F2 - insight into the structural factors for substrate specificity.
Febs J., 283, 2016
|
|
3WRZ
 
 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (without soaking) | | Descriptor: | Beta-lactamase, CALCIUM ION, CHLORIDE ION | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6YEN
 
 | | Crystal structure of AmpC from E. coli with Taniborbactam (VNRX-5133) | | Descriptor: | (10aR)-2-(((1r,4R)-4-((2-aminoethyl)amino)cyclohexyl)methyl)-6-carboxy-4-hydroxy-4,10a-dihydro-10H-benzo[5,6][1,2]oxaborinino[2,3-b][1,4,2]oxazaborol-4-uide, (3~{R})-3-[2-[4-(2-azanylethylamino)cyclohexyl]ethanoylamino]-2-oxidanyl-3,4-dihydro-1,2-benzoxaborinine-8-carboxylic acid, 1,2-ETHANEDIOL, ... | | Authors: | Lang, P.A, Brem, J, Schofield, C.J. | | Deposit date: | 2020-03-25 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.418 Å) | | Cite: | Bicyclic Boronates as Potent Inhibitors of AmpC, the Class C beta-Lactamase from Escherichia coli .
Biomolecules, 10, 2020
|
|
6YPD
 
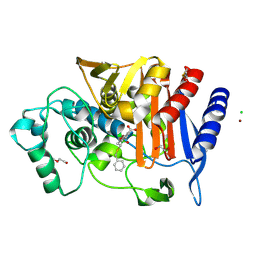 | | Crystal structure of AmpC from E. coli with Cyclic Boronate 3 (CB3 / APC308) | | Descriptor: | (3~{S})-2,2-bis(oxidanyl)-3-(phenylmethylsulfanyl)-3,4-dihydro-1,2-benzoxaborinin-2-ium-8-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase, ... | | Authors: | Lang, P.A, Brem, J, Schofield, C.J. | | Deposit date: | 2020-04-15 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Bicyclic Boronates as Potent Inhibitors of AmpC, the Class C beta-Lactamase from Escherichia coli .
Biomolecules, 10, 2020
|
|
6YEO
 
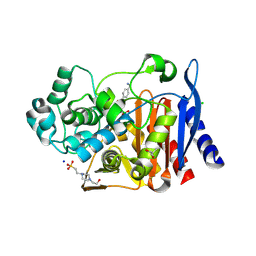 | | Crystal structure of AmpC from E. coli with cyclic boronate 2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (4~{R})-4-[[4-(aminomethyl)phenyl]carbonylamino]-3,3-bis(oxidanyl)-2-oxa-3-boranuidabicyclo[4.4.0]deca-1(10),6,8-triene-10-carboxylic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Lang, P.A, Schofield, C.J, Brem, J. | | Deposit date: | 2020-03-25 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.037 Å) | | Cite: | Bicyclic Boronates as Potent Inhibitors of AmpC, the Class C beta-Lactamase from Escherichia coli .
Biomolecules, 10, 2020
|
|
4QD4
 
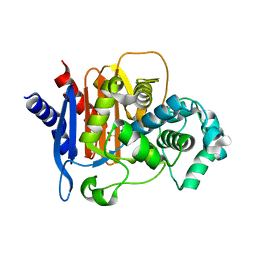 | | Structure of ADC-68, a Novel Carbapenem-Hydrolyzing Class C Extended-Spectrum -Lactamase from Acinetobacter baumannii | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-lactamase ADC-68, CITRIC ACID | | Authors: | Hong, M.K, Kang, L.W. | | Deposit date: | 2014-05-13 | | Release date: | 2015-01-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of ADC-68, a novel carbapenem-hydrolyzing class C extended-spectrum beta-lactamase isolated from Acinetobacter baumannii
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8AIK
 
 | |
8AJI
 
 | | Crystal structure of DltE from L. plantarum, TCEP form | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, Beta-lactamase family protein, GLYCEROL, ... | | Authors: | Ravaud, S, Nikolopoulos, N, Grangeasse, C. | | Deposit date: | 2022-07-28 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure-function analysis of Lactiplantibacillus plantarum DltE& reveals D-alanylated lipoteichoic acids as direct cues supporting Drosophila juvenile growth.
Elife, 12, 2023
|
|
8AKH
 
 | | Crystal structure of DltE from L. plantarum soaked with LTA | | Descriptor: | Beta-lactamase family protein, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Ravaud, S, Nikolopoulos, N, Grangeasse, C. | | Deposit date: | 2022-07-29 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-function analysis of Lactiplantibacillus plantarum DltE& reveals D-alanylated lipoteichoic acids as direct cues supporting Drosophila juvenile growth.
Elife, 12, 2023
|
|
8AGR
 
 | | Crystal structure of DltE from L. plantarum, apo form | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase family protein, SULFATE ION | | Authors: | Ravaud, S, Nikolopoulos, N, Grangeasse, C. | | Deposit date: | 2022-07-20 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure-function analysis of Lactiplantibacillus plantarum DltE& reveals D-alanylated lipoteichoic acids as direct cues supporting Drosophila juvenile growth.
Elife, 12, 2023
|
|
4U0X
 
 | | Structure of ADC-7 beta-lactamase in complex with boronic acid inhibitor S02030 | | Descriptor: | 1-{(2R)-2-(dihydroxyboranyl)-2-[(thiophen-2-ylacetyl)amino]ethyl}-1H-1,2,3-triazole-4-carboxylic acid, ADC-7 beta-lactamase, PHOSPHATE ION | | Authors: | Powers, R.A, Wallar, B.J, Swanson, H.C. | | Deposit date: | 2014-07-14 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Biochemical and Structural Analysis of Inhibitors Targeting the ADC-7 Cephalosporinase of Acinetobacter baumannii.
Biochemistry, 53, 2014
|
|
4U0T
 
 | | Crystal structure of ADC-7 beta-lactamase | | Descriptor: | ADC-7 beta-lactamase, PHOSPHATE ION | | Authors: | Powers, R.A, Wallar, B.J. | | Deposit date: | 2014-07-14 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Biochemical and Structural Analysis of Inhibitors Targeting the ADC-7 Cephalosporinase of Acinetobacter baumannii.
Biochemistry, 53, 2014
|
|
5CHM
 
 | | CRYSTAL STRUCTURE OF Fox-4 cephamycinase complexed with ceftazidime BATSI (LP06) | | Descriptor: | ACETATE ION, Beta-lactamase, PINACOL[[2-AMINO-ALPHA-(1-CARBOXY-1-METHYLETHOXYIMINO)-4-THIAZOLEACETYL]AMINO]METHANEBORONATE, ... | | Authors: | Malashkevich, V.N, Toro, R, Lefurgy, S, Almo, S.C. | | Deposit date: | 2015-07-10 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of FOX-4 Cephamycinase in Complex with Transition-State Analog Inhibitors.
Biomolecules, 10, 2020
|
|
