3OYH
 
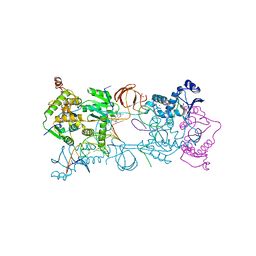 | | Crystal structure of the Prototype Foamy Virus (PFV) intasome in complex with magnesium and the INSTI MK0536 | | Descriptor: | 6-(3-chloro-4-fluorobenzyl)-4-hydroxy-N,N-dimethyl-2-(1-methylethyl)-3,5-dioxo-2,3,5,6,7,8-hexahydro-2,6-naphthyridine-1-carboxamide, AMMONIUM ION, DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), ... | | Authors: | Hare, S, Cherepanov, P. | | Deposit date: | 2010-09-23 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OY9
 
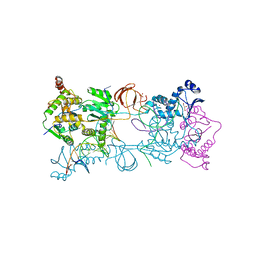 | |
3OYM
 
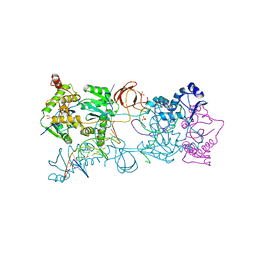 | | Crystal structure of the PFV N224H mutant intasome bound to manganese | | Descriptor: | AMMONIUM ION, DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*GP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP*AP*CP*A)-3'), ... | | Authors: | Hare, S, Cherepanov, P. | | Deposit date: | 2010-09-23 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OYA
 
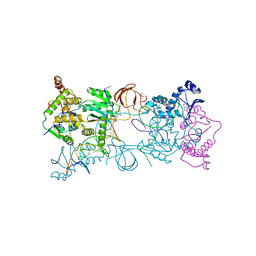 | |
3S3N
 
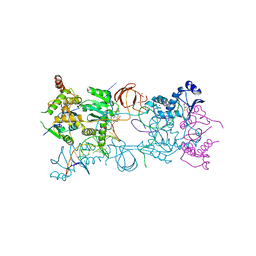 | | Crystal structure of the Prototype Foamy Virus (PFV) S217H mutant intasome in complex with magnesium and Dolutegravir (S/GSK1349572) | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, 5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3', 5'-D(*TP*GP*CP*GP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP*AP*CP*A)-3', ... | | Authors: | Hare, S, Cherepanov, P. | | Deposit date: | 2011-05-18 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural and Functional Analyses of the Second-Generation Integrase Strand Transfer Inhibitor Dolutegravir (S/GSK1349572).
Mol.Pharmacol., 80, 2011
|
|
4BE2
 
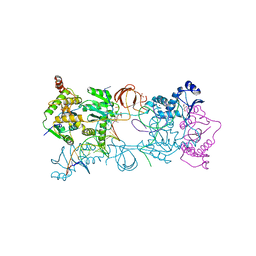 | | PFV intasome with inhibitor XZ-259 | | Descriptor: | 17 NUCLEOTIDE PREPROCESSED PFV DONOR DNA (TRANSFERRED STRAND) *AP*CP*A)-3', 19 NUCLEOTIDE PREPROCESSED PFV DONOR DNA (NON-TRANSFERRED STRAND), 2-(3-chloro-4-fluorobenzyl)-6,7-dihydroxy-N,N-dimethyl-1-oxo-2,3-dihydro-1H-isoindole-4-sulfonamide, ... | | Authors: | Hare, S, Cherepanov, P. | | Deposit date: | 2012-10-08 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Activities, Crystal Structures and Molecular Dynamics of Dihydro-1H-Isoindole Derivatives, Inhibitors of HIV-1 Integrase.
Acs Chem.Biol., 8, 2013
|
|
4BE1
 
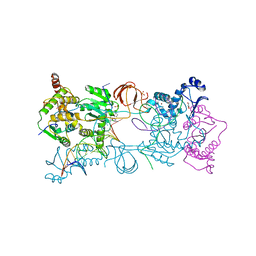 | | PFV intasome with inhibitor XZ-116 | | Descriptor: | 17 NUCLEOTIDE PREPROCESSED PFV DONOR DNA (TRANSFERRED STRAND), 19 NUCLEOTIDE PREPROCESSED PFV DONOR DNA (NON-TRANSFERRED STRAND), 2-(3-chloro-2-fluorobenzyl)-6,7-dihydroxy-2,3-dihydro-1H-isoindol-1-one, ... | | Authors: | Hare, S, Cherepanov, P. | | Deposit date: | 2012-10-08 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Activities, Crystal Structures and Molecular Dynamics of Dihydro-1H-Isoindole Derivatives, Inhibitors of HIV-1 Integrase.
Acs Chem.Biol., 8, 2013
|
|
4BAC
 
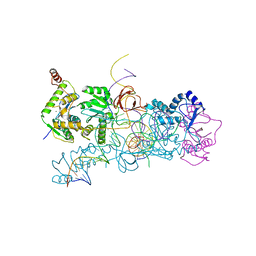 | | prototype foamy virus strand transfer complexes on product DNA | | Descriptor: | 5'-D(*AP*GP*GP*AP*GP*CP*CP*AP*AP*GP*AP*CP*GP*GP *AP*TP*CP)-3', 5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP *TP*TP*GP*TP*AP)-3', DNA (38-MER), ... | | Authors: | Yin, Z, Lapkouski, M, Yang, W, Craigie, R. | | Deposit date: | 2012-09-12 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.263 Å) | | Cite: | Assembly of Prototype Foamy Virus Strand Transfer Complexes on Product DNA Bypassing Catalysis of Integration.
Protein Sci., 21, 2012
|
|
4BE0
 
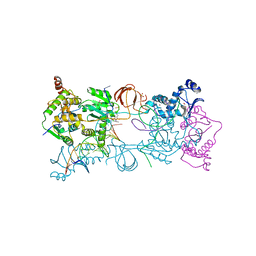 | | PFV intasome with inhibitor XZ-115 | | Descriptor: | 17 NUCLEOTIDE PREPROCESSED PFV DONOR DNA (TRANSFERRED STRAND), 19 NUCLEOTIDE PREPROCESSED PFV DONOR DNA (NON-TRANSFERRED STRAND), 2-(3-chloro-2-fluorobenzyl)-4,5-dihydroxy-1H-isoindole-1,3(2H)-dione, ... | | Authors: | Hare, S, Cherepanov, P. | | Deposit date: | 2012-10-08 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Activities, Crystal Structures and Molecular Dynamics of Dihydro-1H-Isoindole Derivatives, Inhibitors of HIV-1 Integrase.
Acs Chem.Biol., 8, 2013
|
|
4BDY
 
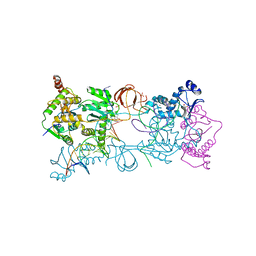 | | PFV intasome with inhibitor XZ-89 | | Descriptor: | 17 NUCLEOTIDE PREPROCESSED PFV DONOR DNA (TRANSFERRED STRAND), 19 NUCLEOTIDE PREPROCESSED PFV DONOR DNA (NON-TRANSFERRED STRAND), 2-(3-chloro-4-fluorobenzyl)-4,5-dihydroxy-1H-isoindole-1,3(2H)-dione, ... | | Authors: | Hare, S, Cherepanov, P. | | Deposit date: | 2012-10-08 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Activities, Crystal Structures and Molecular Dynamics of Dihydro-1H-Isoindole Derivatives, Inhibitors of HIV-1 Integrase.
Acs Chem.Biol., 8, 2013
|
|
4BDZ
 
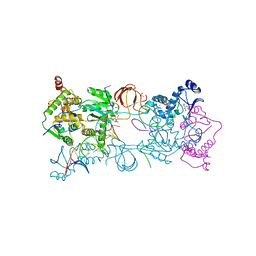 | | PFV intasome with inhibitor XZ-90 | | Descriptor: | 17 NUCLEOTIDE PREPROCESSED PFV DONOR DNA (TRANSFERRED STRAND), 19 NUCLEOTIDE PREPROCESSED PFV DONOR DNA (NON-TRANSFERRED STRAND), 2-[(3-chloranyl-4-fluoranyl-phenyl)methyl]-6,7-bis(oxidanyl)isoindol-1-one, ... | | Authors: | Hare, S, Cherepanov, P. | | Deposit date: | 2012-10-08 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Activities, Crystal Structures and Molecular Dynamics of Dihydro-1H-Isoindole Derivatives, Inhibitors of HIV-1 Integrase.
Acs Chem.Biol., 8, 2013
|
|
4E7I
 
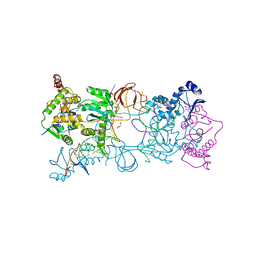 | | PFV intasome freeze-trapped prior to 3'-processing, Mn-bound form (UI-Mn) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*GP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP*AP*CP*AP*AP*T)-3'), ... | | Authors: | Hare, S, Cherepanov, P. | | Deposit date: | 2012-03-17 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5301 Å) | | Cite: | 3'-Processing and strand transfer catalysed by retroviral integrase in crystallo.
Embo J., 31, 2012
|
|
7SEP
 
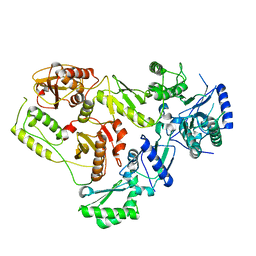 | |
8E14
 
 | | Cryo-EM structure of Rous sarcoma virus strand transfer complex | | Descriptor: | DNA (42-MER), DNA (5'-D(*AP*AP*TP*GP*TP*TP*GP*TP*CP*TP*TP*AP*TP*GP*CP*AP*AP*TP*AP*CP*TP*C)-3'), DNA (5'-D(*AP*GP*TP*GP*TP*CP*TP*TP*CP*TP*TP*CP*TP*TP*TP*C)-3'), ... | | Authors: | Pandey, K.K, Bera, S, Shi, K, Aihara, H, Grandgenett, D.P. | | Deposit date: | 2022-08-09 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Molecular determinants for Rous sarcoma virus intasome assemblies involved in retroviral integration.
J.Biol.Chem., 299, 2023
|
|
8W2R
 
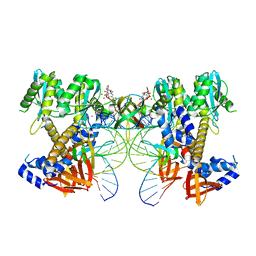 | | HIV-1 P5-IN intasome core | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (5'-D(*AP*CP*TP*GP*CP*TP*AP*GP*AP*GP*AP*TP*TP*TP*TP*CP*CP*CP*G)-3'), DNA (5'-D(P*CP*GP*GP*GP*AP*AP*AP*AP*TP*CP*TP*CP*TP*AP*GP*CP*A)-3'), ... | | Authors: | Li, M, Craigie, R. | | Deposit date: | 2024-02-21 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | HIV-1 Integrase Assembles Multiple Species of Stable Synaptic Complex Intasomes That Are Active for Concerted DNA Integration In vitro.
J.Mol.Biol., 436, 2024
|
|
8W34
 
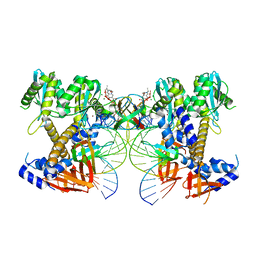 | | HIV-1 intasome core assembled with wild-type integrase, 1F | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (5'-D(*AP*CP*TP*GP*CP*TP*AP*GP*AP*GP*AP*TP*TP*TP*TP*CP*CP*AP*C)-3'), DNA (5'-D(P*GP*TP*GP*GP*AP*AP*AP*AP*TP*CP*TP*CP*TP*AP*GP*CP*A)-3'), ... | | Authors: | Li, M, Craigie, R. | | Deposit date: | 2024-02-21 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | HIV-1 Integrase Assembles Multiple Species of Stable Synaptic Complex Intasomes That Are Active for Concerted DNA Integration In vitro.
J.Mol.Biol., 436, 2024
|
|
8W09
 
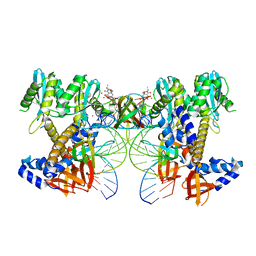 | | HIV-1 wild-type intasome core | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, Integrase, MAGNESIUM ION, ... | | Authors: | Li, M, Craigie, R. | | Deposit date: | 2024-02-13 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | HIV-1 Integrase Assembles Multiple Species of Stable Synaptic Complex Intasomes That Are Active for Concerted DNA Integration In vitro.
J.Mol.Biol., 436, 2024
|
|
7PEL
 
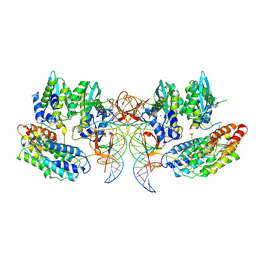 | | CryoEM structure of simian T-cell lymphotropic virus intasome in complex with PP2A regulatory subunit B56 gamma | | Descriptor: | DNA (5'-D(*AP*CP*TP*GP*TP*GP*TP*TP*TP*GP*GP*CP*GP*CP*TP*TP*CP*TP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*AP*GP*AP*AP*GP*CP*GP*CP*CP*AP*AP*AP*CP*AP*CP*A)-3'), Isoform 3 of PC4 and SFRS1-interacting protein,Isoform Gamma-2 of Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform, ... | | Authors: | Barski, M, Pye, V.E, Nans, A, Cherepanov, P, Maertens, G.N. | | Deposit date: | 2021-08-10 | | Release date: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Cryo-EM structure of the deltaretroviral intasome in complex with the PP2A regulatory subunit B56gamma.
Nat Commun, 11, 2020
|
|
7Q5B
 
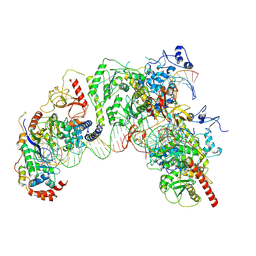 | | Cryo-EM structure of Ty3 retrotransposon targeting a TFIIIB-bound tRNA gene | | Descriptor: | DNA (19-MER), DNA (31-MER), DNA (34-MER), ... | | Authors: | Abascal-Palacios, G, Jochem, L, Pla-Prats, C, Beuron, F, Vannini, A. | | Deposit date: | 2021-11-03 | | Release date: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Structural basis of Ty3 retrotransposon integration at RNA Polymerase III-transcribed genes.
Nat Commun, 12, 2021
|
|
1EX4
 
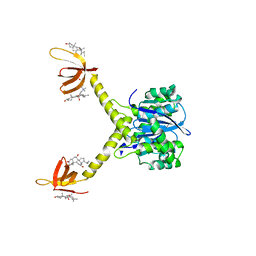 | | HIV-1 INTEGRASE CATALYTIC CORE AND C-TERMINAL DOMAIN | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, INTEGRASE | | Authors: | Chen, J.C.-H, Krucinski, J, Miercke, L.J.W, Finer-Moore, J.S, Tang, A.H, Leavitt, A.D, Stroud, R.M. | | Deposit date: | 2000-04-28 | | Release date: | 2000-06-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
7OUF
 
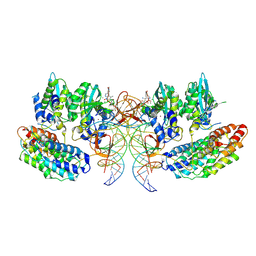 | | Structure of the STLV intasome:B56 complex bound to the strand-transfer inhibitor XZ450 | | Descriptor: | 4-azanyl-~{N}-[[2,4-bis(fluoranyl)phenyl]methyl]-6-[3-(dimethylamino)-3-oxidanylidene-propyl]-1-oxidanyl-2-oxidanylidene-1,8-naphthyridine-3-carboxamide, DNA (5'-D(*AP*CP*TP*GP*TP*GP*TP*TP*TP*GP*GP*CP*GP*CP*TP*TP*CP*TP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*AP*GP*AP*AP*GP*CP*GP*CP*CP*AP*AP*AP*CP*AP*CP*A)-3'), ... | | Authors: | Barski, M.S, Ballandras-Colas, A, Cronin, N.B, Pye, V.E, Cherepanov, P, Maertens, G.N. | | Deposit date: | 2021-06-11 | | Release date: | 2021-08-18 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for the inhibition of HTLV-1 integration inferred from cryo-EM deltaretroviral intasome structures.
Nat Commun, 12, 2021
|
|
7OUG
 
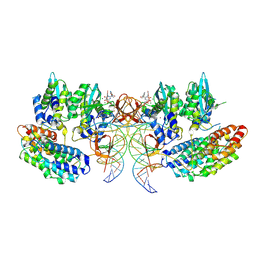 | | STLV-1 intasome:B56 in complex with the strand-transfer inhibitor raltegravir | | Descriptor: | DNA (5'-D(*AP*CP*TP*GP*TP*GP*TP*TP*TP*GP*GP*CP*GP*CP*TP*TP*CP*TP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*AP*GP*AP*AP*GP*CP*GP*CP*CP*AP*AP*AP*CP*AP*CP*A)-3'), Integrase, ... | | Authors: | Barski, M.S, Ballandras-Colas, A, Cronin, N.B, Pye, V.E, Cherepanov, P, Maertens, G.N. | | Deposit date: | 2021-06-11 | | Release date: | 2021-08-18 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for the inhibition of HTLV-1 integration inferred from cryo-EM deltaretroviral intasome structures.
Nat Commun, 12, 2021
|
|
7OUH
 
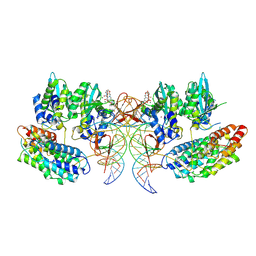 | | Structure of the STLV intasome:B56 complex bound to the strand-transfer inhibitor bictegravir | | Descriptor: | Bictegravir, DNA (5'-D(*AP*CP*TP*GP*TP*GP*TP*TP*TP*GP*GP*CP*GP*CP*TP*TP*CP*TP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*AP*GP*AP*AP*GP*CP*GP*CP*CP*AP*AP*AP*CP*AP*CP*A)-3'), ... | | Authors: | Barski, M.S, Ballandras-Colas, A, Cronin, N.B, Pye, V.E, Cherepanov, P, Maertens, G.N. | | Deposit date: | 2021-06-11 | | Release date: | 2021-08-18 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for the inhibition of HTLV-1 integration inferred from cryo-EM deltaretroviral intasome structures.
Nat Commun, 12, 2021
|
|
5U1C
 
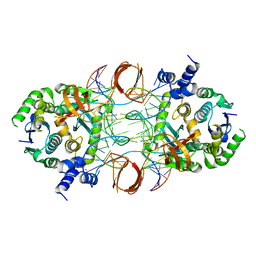 | |
3JCA
 
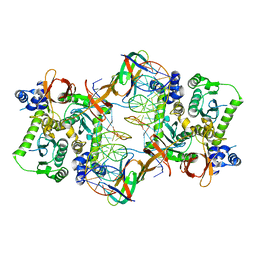 | | Core model of the Mouse Mammary Tumor Virus intasome | | Descriptor: | 5'-D(*AP*AP*TP*GP*CP*CP*GP*CP*AP*GP*TP*CP*GP*GP*CP*CP*GP*AP*CP*CP*TP*G)-3', 5'-D(*CP*AP*GP*GP*TP*CP*GP*GP*CP*CP*GP*AP*CP*TP*GP*CP*GP*GP*CP*A)-3', Integrase, ... | | Authors: | Lyumkis, D.L, Ballandras-Colas, A, Brown, M, Cook, N.J, Dewdney, T.G, Demeler, B, Cherepanov, P, Engelman, A.N. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-17 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryo-EM reveals a novel octameric integrase structure for betaretroviral intasome function.
Nature, 530, 2016
|
|
