4CI8
 
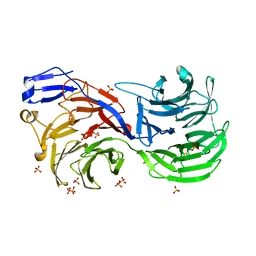 | |
5A9Q
 
 | | Human nuclear pore complex | | 分子名称: | NUCLEAR PORE COMPLEX PROTEIN NUP107, NUCLEAR PORE COMPLEX PROTEIN NUP133, NUCLEAR PORE COMPLEX PROTEIN NUP155, ... | | 著者 | von Appen, A, Kosinski, J, Sparks, L, Ori, A, DiGuilio, A, Vollmer, B, Mackmull, M, Banterle, N, Parca, L, Kastritis, P, Buczak, K, Mosalaganti, S, Hagen, W, Andres-Pons, A, Lemke, E.A, Bork, P, Antonin, W, Glavy, J.S, Bui, K.H, Beck, M. | | 登録日 | 2015-07-22 | | 公開日 | 2015-09-30 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (23 Å) | | 主引用文献 | In Situ Structural Analysis of the Human Nuclear Pore Complex
Nature, 526, 2015
|
|
4BL0
 
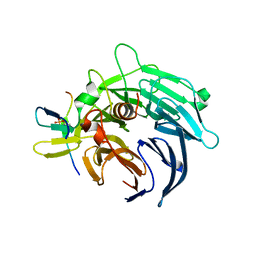 | | Crystal structure of yeast Bub3-Bub1 bound to phospho-Spc105 | | 分子名称: | CELL CYCLE ARREST PROTEIN BUB3, CHECKPOINT SERINE/THREONINE-PROTEIN KINASE BUB1, MAGNESIUM ION, ... | | 著者 | Primorac, I, Weir, J.R, Musacchio, A. | | 登録日 | 2013-04-30 | | 公開日 | 2013-09-18 | | 最終更新日 | 2013-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Bub3 Reads Phosphorylated Melt Repeats to Promote Spindle Assembly Checkpoint Signaling
Elife, 2, 2013
|
|
4E5Z
 
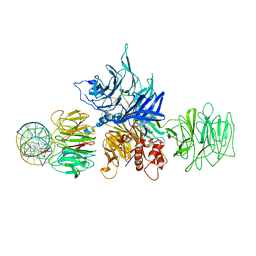 | |
5BJS
 
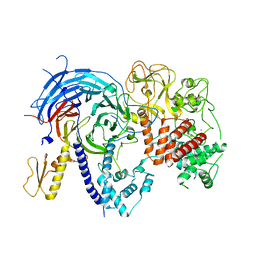 | | Apo ctPRC2 in an autoinhibited state | | 分子名称: | Histone-lysine N-methyltransferase EZH2, Polycomb protein SUZ12, Polycomb Protein EED, ... | | 著者 | Bratkowski, M.A, Liu, X. | | 登録日 | 2016-10-22 | | 公開日 | 2017-06-14 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.189 Å) | | 主引用文献 | Polycomb repressive complex 2 in an autoinhibited state.
J. Biol. Chem., 292, 2017
|
|
4E54
 
 | |
5C9Z
 
 | |
5CXC
 
 | |
5CYK
 
 | | Structure of Ytm1 bound to the C-terminal domain of Erb1-R486E | | 分子名称: | CHLORIDE ION, Ribosome biogenesis protein ERB1, Ribosome biogenesis protein YTM1 | | 著者 | Wegrecki, M, Bravo, J. | | 登録日 | 2015-07-30 | | 公開日 | 2015-10-28 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The structure of Erb1-Ytm1 complex reveals the functional importance of a high-affinity binding between two beta-propellers during the assembly of large ribosomal subunits in eukaryotes.
Nucleic Acids Res., 43, 2015
|
|
5EM2
 
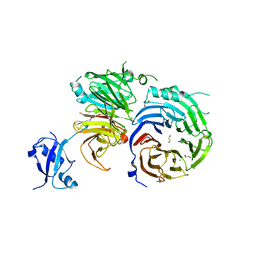 | | Crystal structure of the Erb1-Ytm1 complex | | 分子名称: | 1,2-ETHANEDIOL, MAGNESIUM ION, Ribosome biogenesis protein ERB1, ... | | 著者 | Ahmed, Y.L, Sinning, I. | | 登録日 | 2015-11-05 | | 公開日 | 2015-12-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Concerted removal of the Erb1-Ytm1 complex in ribosome biogenesis relies on an elaborate interface.
Nucleic Acids Res., 44, 2016
|
|
5EMM
 
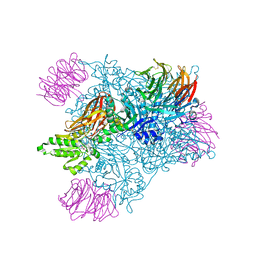 | |
5CVL
 
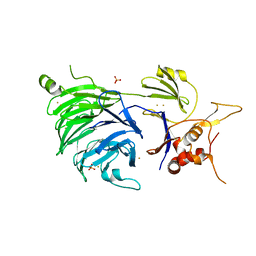 | | WDR48 (UAF-1), residues 2-580 | | 分子名称: | GOLD ION, PHOSPHATE ION, WD repeat-containing protein 48 | | 著者 | HARRIS, S.F, YIN, J. | | 登録日 | 2015-07-27 | | 公開日 | 2015-10-07 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural Insights into WD-Repeat 48 Activation of Ubiquitin-Specific Protease 46.
Structure, 23, 2015
|
|
5EML
 
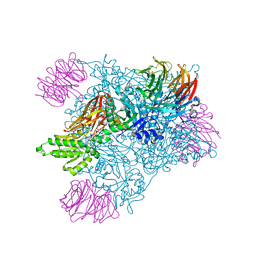 | |
5EMK
 
 | |
5CXB
 
 | |
5EMJ
 
 | |
3W15
 
 | | Structure of peroxisomal targeting signal 2 (PTS2) of Saccharomyces cerevisiae 3-ketoacyl-CoA thiolase in complex with Pex7p and Pex21p | | 分子名称: | 3-ketoacyl-CoA thiolase, peroxisomal, Maltose-binding periplasmic protein, ... | | 著者 | Pan, D, Nakatsu, T, Kato, H. | | 登録日 | 2012-11-06 | | 公開日 | 2013-07-03 | | 最終更新日 | 2017-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of peroxisomal targeting signal-2 bound to its receptor complex Pex7p-Pex21p
Nat.Struct.Mol.Biol., 20, 2013
|
|
3V7D
 
 | | Crystal Structure of ScSkp1-ScCdc4-pSic1 peptide complex | | 分子名称: | Cell division control protein 4, Protein SIC1, Suppressor of kinetochore protein 1 | | 著者 | Tang, X, Orlicky, S, Mittag, T, Csizmok, V, Pawson, T, Forman-Kay, J, Sicheri, F, Tyers, M. | | 登録日 | 2011-12-20 | | 公開日 | 2012-05-02 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.306 Å) | | 主引用文献 | Composite low affinity interactions dictate recognition of the cyclin-dependent kinase inhibitor Sic1 by the SCFCdc4 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AEZ
 
 | | Crystal Structure of Mitotic Checkpoint Complex | | 分子名称: | MITOTIC SPINDLE CHECKPOINT COMPONENT MAD2, MITOTIC SPINDLE CHECKPOINT COMPONENT MAD3, WD REPEAT-CONTAINING PROTEIN SLP1 | | 著者 | Kulkarni, K.A, Chao, W.C.H, Zhang, Z, Barford, D. | | 登録日 | 2012-01-13 | | 公開日 | 2012-03-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of the Mitotic Checkpoint Complex
Nature, 484, 2012
|
|
4A0B
 
 | | Structure of hsDDB1-drDDB2 bound to a 16 bp CPD-duplex (pyrimidine at D-1 position) at 3.8 A resolution (CPD 4) | | 分子名称: | 5'-D(*CP*CP*TP*GP*CP*TP*CP*CP*TP*TP*TP*CP*AP*CP*CP*C)-3', 5'-D(*DGP*GP*TP*GP*AP*AP*AP*(TTD)P*AP*GP*CP*AP*GP*DGP)-3', DNA DAMAGE-BINDING PROTEIN 1, ... | | 著者 | Scrima, A, Fischer, E.S, Iwai, S, Gut, H, Thoma, N.H. | | 登録日 | 2011-09-08 | | 公開日 | 2011-11-30 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation
Cell(Cambridge,Mass.), 147, 2011
|
|
4A0L
 
 | | Structure of DDB1-DDB2-CUL4B-RBX1 bound to a 12 bp abasic site containing DNA-duplex | | 分子名称: | 12 BP DNA DUPLEX, 12 BP THF CONTAINING DNA DUPLEX, CULLIN-4B, ... | | 著者 | Fischer, E.S, Scrima, A, Gut, H, Thoma, N.H. | | 登録日 | 2011-09-09 | | 公開日 | 2011-12-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (7.4 Å) | | 主引用文献 | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation.
Cell(Cambridge,Mass.), 147, 2011
|
|
4A0A
 
 | | Structure of hsDDB1-drDDB2 bound to a 16 bp CPD-duplex (pyrimidine at D-1 position) at 3.6 A resolution (CPD 3) | | 分子名称: | 5'-D(*CP*CP*TP*GP*CP*TP*CP*CP*TP*TP*TP*CP*AP*CP*CP*C)-3', 5'-D(*GP*GP*TP*GP*AP*AP*AP*(TTD)P*AP*GP*CP*AP*GP*DGP)-3', CALCIUM ION, ... | | 著者 | Scrima, A, Fischer, E.S, Iwai, S, Gut, H, Thoma, N.H. | | 登録日 | 2011-09-08 | | 公開日 | 2011-11-30 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation
Cell(Cambridge,Mass.), 147, 2011
|
|
4A0K
 
 | | STRUCTURE OF DDB1-DDB2-CUL4A-RBX1 BOUND TO A 12 BP ABASIC SITE CONTAINING DNA-DUPLEX | | 分子名称: | 12 BP DNA, 12 BP THF CONTAINING DNA, CULLIN-4A, ... | | 著者 | Fischer, E.S, Scrima, A, Gut, H, Thoma, N.H. | | 登録日 | 2011-09-09 | | 公開日 | 2011-12-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (5.93 Å) | | 主引用文献 | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation.
Cell(Cambridge,Mass.), 147, 2011
|
|
4A08
 
 | | Structure of hsDDB1-drDDB2 bound to a 13 bp CPD-duplex (purine at D-1 position) at 3.0 A resolution (CPD 1) | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-D(*AP*CP*GP*CP*GP*AP*(TTD)P*GP*CP*GP*CP*CP*C)-3', 5'-D(*TP*GP*GP*GP*CP*GP*CP*CP*CP*TP*CP*GP*CP*G)-3', ... | | 著者 | Scrima, A, Fischer, E.S, Iwai, S, Gut, H, Thoma, N.H. | | 登録日 | 2011-09-08 | | 公開日 | 2011-11-30 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation
Cell(Cambridge,Mass.), 147, 2011
|
|
4A09
 
 | | Structure of hsDDB1-drDDB2 bound to a 15 bp CPD-duplex (purine at D-1 position) at 3.1 A resolution (CPD 2) | | 分子名称: | 5'-D(*CP*CP*TP*GP*CP*TP*CP*CP*TP*TP*TP*CP*AP*CP*CP*C)-3', 5'-D(*GP*GP*TP*GP*AP*AP*AP*(TTD)P*AP*GP*CP*AP*GP*GP)-3', CALCIUM ION, ... | | 著者 | Scrima, A, Fischer, E.S, Iwai, S, Gut, H, Thoma, N.H. | | 登録日 | 2011-09-08 | | 公開日 | 2011-11-30 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation
Cell(Cambridge,Mass.), 147, 2011
|
|
