4YSI
 
 | | Structure of USP7 with a novel viral protein | | Descriptor: | GLYCEROL, SER-PRO-GLY-GLU-GLY-PRO-SER-GLY, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Chavoshi, S, Saridakis, V. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Structure of USP7 with a novel viral protein
J.Biol.Chem., 2016
|
|
4Z96
 
 | | Crystal structure of DNMT1 in complex with USP7 | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Zhang, Z.M, Song, J. | | Deposit date: | 2015-04-09 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of DNMT1 in complex with USP7
To Be Published
|
|
4Z97
 
 | |
5VSB
 
 | | Structure of DUB complex | | Descriptor: | 7-chloro-3-{[4-hydroxy-1-(3-phenylpropanoyl)piperidin-4-yl]methyl}quinazolin-4(3H)-one, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2017-05-11 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Guided Development of a Potent and Selective Non-covalent Active-Site Inhibitor of USP7.
Cell Chem Biol, 24, 2017
|
|
5VS6
 
 | | Structure of DUB complex | | Descriptor: | ACETATE ION, GLYCEROL, N-[3-({4-hydroxy-1-[(3R)-3-phenylbutanoyl]piperidin-4-yl}methyl)-4-oxo-3,4-dihydroquinazolin-7-yl]-3-(4-methylpiperazin-1-yl)propanamide, ... | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2017-05-11 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure-Guided Development of a Potent and Selective Non-covalent Active-Site Inhibitor of USP7.
Cell Chem Biol, 24, 2017
|
|
5VSK
 
 | | Structure of DUB complex | | Descriptor: | 7-chloro-3-({4-hydroxy-1-[(3S)-3-phenylbutanoyl]piperidin-4-yl}methyl)quinazolin-4(3H)-one, Ubiquitin carboxyl-terminal hydrolase 7, ZINC ION | | Authors: | Seo, H.-Y, Dhe-Paganon, S. | | Deposit date: | 2017-05-11 | | Release date: | 2017-12-20 | | Last modified: | 2018-01-03 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Structure-Guided Development of a Potent and Selective Non-covalent Active-Site Inhibitor of USP7.
Cell Chem Biol, 24, 2017
|
|
2F1Z
 
 | | Crystal structure of HAUSP | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Hu, M, Gu, L, Jeffrey, P.D, Shi, Y. | | Deposit date: | 2005-11-15 | | Release date: | 2006-02-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis of Competitive Recognition of p53 and MDM2 by HAUSP/USP7: Implications for the Regulation of the p53-MDM2 Pathway.
Plos Biol., 4, 2006
|
|
5VBT
 
 | | Crystal structure of a highly specific and potent USP7 ubiquitin variant inhibitor | | Descriptor: | UBH04 | | Authors: | DONG, A, DONG, X, LIU, L, GUO, Y, LI, Y, ZHANG, W, WALKER, J.R, SIDHU, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, TONG, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-03-30 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structure of a highly specific and potent USP7 ubiquitin variant inhibitor
to be published
|
|
4JJQ
 
 | | Crystal structure of usp7-ntd with an e2 enzyme | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7, Ubiquitin-conjugating enzyme E2 E1 | | Authors: | Saridakis, V. | | Deposit date: | 2013-03-08 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Ubiquitin-specific protease 7 is a regulator of ubiquitin-conjugating enzyme UbE2E1.
J. Biol. Chem., 288, 2013
|
|
5GG4
 
 | | Crystal structure of USP7 with RNF169 peptide | | Descriptor: | Peptide from E3 ubiquitin-protein ligase RNF169, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Jiang, Y, Gong, Q. | | Deposit date: | 2016-06-15 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Dual-utility NLS drives RNF169-dependent DNA damage responses.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4HK2
 
 | | U7Ub25.2540 | | Descriptor: | SULFATE ION, Ubiquitin | | Authors: | Murray, J.M, Rouge, L. | | Deposit date: | 2012-10-14 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Potent and selective inhibitors of USP7/HAUSP activity by protein conformational stabilization
to be published
|
|
4HJK
 
 | | U7Ub7 Disulfide variant | | Descriptor: | PHOSPHATE ION, Ubiquitin | | Authors: | Murray, J.M, Rouge, L. | | Deposit date: | 2012-10-12 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.784 Å) | | Cite: | Potent and selective inhibitors of USP7/HAUSP activity by protein conformational stabilization
to be published
|
|
6P5L
 
 | | Crystal Structure of Ubl123 with an EZH2 peptide | | Descriptor: | PRO-ARG-LYS-LYS-LYS-ARG-LYS-HIS, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Saridakis, V. | | Deposit date: | 2019-05-30 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.296 Å) | | Cite: | Structural Basis of the Interaction Between Ubiquitin Specific Protease 7 and Enhancer of Zeste Homolog 2.
J.Mol.Biol., 432, 2020
|
|
6GDJ
 
 | |
1NB8
 
 | | Structure of the catalytic domain of USP7 (HAUSP) | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Hu, M, Li, P, Li, M, Li, W, Yao, T, Wu, J.-W, Gu, W, Cohen, R.E, Shi, Y. | | Deposit date: | 2002-12-02 | | Release date: | 2003-01-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde
Cell(Cambridge,Mass.), 111, 2002
|
|
5TOG
 
 | | Room temperature structure of ubiquitin variant u7ub25.2540 | | Descriptor: | Polyubiquitin-B, SULFATE ION | | Authors: | Biel, J.T, Thompson, M.C, Cunningham, C.N, Corn, J.E, Fraser, J.S. | | Deposit date: | 2016-10-17 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Flexibility and Design: Conformational Heterogeneity along the Evolutionary Trajectory of a Redesigned Ubiquitin.
Structure, 25, 2017
|
|
5TOF
 
 | | Room temperature structure of ubiquitin variant u7ub25 | | Descriptor: | Polyubiquitin-B, SULFATE ION | | Authors: | Biel, J.T, Thompson, M.C, Cunningham, C.N, Corn, J.E, Fraser, J.S. | | Deposit date: | 2016-10-17 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Flexibility and Design: Conformational Heterogeneity along the Evolutionary Trajectory of a Redesigned Ubiquitin.
Structure, 25, 2017
|
|
4Y2E
 
 | | Crystal structure of the catalytic domain of human dual-specificity phosphatase 7 (C232S) | | Descriptor: | Dual specificity protein phosphatase 7, PHOSPHATE ION | | Authors: | Lountos, G.T, Austin, B.P, Tropea, J.E, Waugh, D.S. | | Deposit date: | 2015-02-09 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure of human dual-specificity phosphatase 7, a potential cancer drug target.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
8ITP
 
 | |
8ITN
 
 | | Crystal structure of USP47apo catalytic domain | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 47, ZINC ION | | Authors: | Kim, E.E, Shin, S.C. | | Deposit date: | 2023-03-22 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional characterization of USP47 reveals a hot spot for inhibitor design.
Commun Biol, 6, 2023
|
|
8QH5
 
 | | CryoEM structure of UVSSA(VHS)-CSA-DDB1-DDA1 | | Descriptor: | DET1- and DDB1-associated protein 1, DNA damage-binding protein 1, DNA excision repair protein ERCC-8, ... | | Authors: | Lee, S.-H, Sixma, T.K. | | Deposit date: | 2023-09-06 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | CryoEM structure of UVSSA(VHS)-CSA-DDB1-DDA1
To Be Published
|
|
2XXN
 
 | | Structure of the vIRF4-HAUSP TRAF domain complex | | Descriptor: | K10, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE 7 | | Authors: | Choi, W.C, Hwang, J, Kim, M.H. | | Deposit date: | 2010-11-11 | | Release date: | 2011-11-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Bilateral Inhibition of Hausp Deubiquitinase by a Viral Interferon Regulatory Factor Protein
Nat.Struct.Mol.Biol., 18, 2011
|
|
6GHA
 
 | | USP15 catalytic domain structure | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 15,Ubiquitin carboxyl-terminal hydrolase 15, ZINC ION | | Authors: | Ward, S.J, Gratton, H.E, Caulton, S.G, Emsley, J, Dreveny, I. | | Deposit date: | 2018-05-06 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The structure of the deubiquitinase USP15 reveals a misaligned catalytic triad and an open ubiquitin-binding channel.
J. Biol. Chem., 293, 2018
|
|
6GH9
 
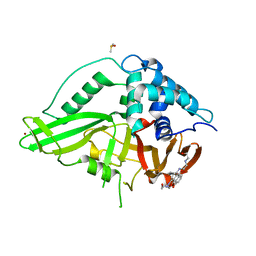 | | USP15 catalytic domain in complex with small molecule | | Descriptor: | 1,4-DIHYDROXY-5,8-BIS({2-[(2-HYDROXYETHYL)AMINO]ETHYL}AMINO)-9,10-ANTHRACENEDIONE, DIMETHYL SULFOXIDE, Ubiquitin carboxyl-terminal hydrolase 15, ... | | Authors: | Ward, S.J, Gratton, H.E, Caulton, S.G, Emsley, J, Dreveny, I. | | Deposit date: | 2018-05-06 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The structure of the deubiquitinase USP15 reveals a misaligned catalytic triad and an open ubiquitin-binding channel.
J. Biol. Chem., 293, 2018
|
|
8HJE
 
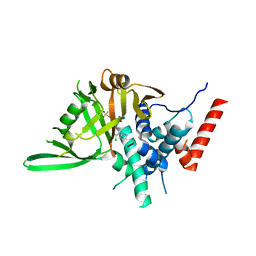 | | Vismodegib binds to the catalytical domain of human Ubiquitin-Specific Protease 28 | | Descriptor: | 2-chloranyl-~{N}-(4-chloranyl-3-pyridin-2-yl-phenyl)-4-methylsulfonyl-benzamide, Ubiquitin carboxyl-terminal hydrolase 28 | | Authors: | Shi, L, Wang, H, Xu, Z, Xiong, B, Zhang, N. | | Deposit date: | 2022-11-23 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure-based discovery of potent USP28 inhibitors derived from Vismodegib.
Eur.J.Med.Chem., 254, 2023
|
|
