4J2X
 
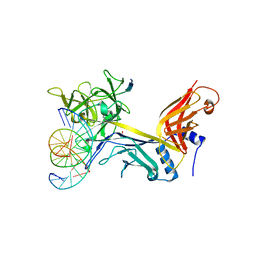 | | CSL (RBP-Jk) with corepressor KyoT2 bound to DNA | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*AP*AP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*AP*GP*T)-3', 5'-D(*TP*TP*AP*CP*TP*GP*TP*GP*GP*GP*AP*AP*AP*GP*A)-3', ... | | Authors: | Collins, K.J, Kovall, R.A. | | Deposit date: | 2013-02-05 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure and Function of the CSL-KyoT2 Corepressor Complex: A Negative Regulator of Notch Signaling.
Structure, 22, 2014
|
|
6IZ4
 
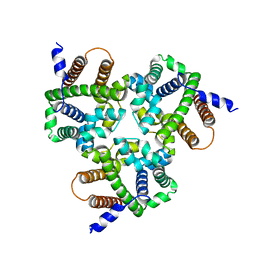 | | Crystal Structure Analysis of TRIC counter-ion channels in calcium release | | Descriptor: | Trimeric intracellular cation channel type B-B | | Authors: | Wang, X.H, Zeng, Y, Gao, F, Su, M, Hendrickson, W.A, Chen, Y.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.098 Å) | | Cite: | Structural basis for activity of TRIC counter-ion channels in calcium release.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7VP5
 
 | |
7VP7
 
 | |
7VP4
 
 | | Structure of a transcription factor and DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*GP*TP*GP*GP*TP*CP*CP*CP*CP*AP*GP*T)-3'), DNA (5'-D(*TP*AP*CP*TP*GP*GP*GP*GP*AP*CP*CP*AP*CP*A)-3'), Transcription factor TCP10 | | Authors: | Zhang, Y, Xu, Y.P, Wang, B, Su, X.D. | | Deposit date: | 2021-10-15 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structural basis for DNA recognition by TCP transcription factors
To Be Published
|
|
7VP1
 
 | |
8IHO
 
 | | Crystal structures of SARS-CoV-2 papain-like protease in complex with covalent inhibitors | | Descriptor: | Papain-like protease nsp3, ZINC ION, covalent inhibitor | | Authors: | Wang, Q, Hu, H, Li, M, Xu, Y. | | Deposit date: | 2023-02-23 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure-Based Design of Potent Peptidomimetic Inhibitors Covalently Targeting SARS-CoV-2 Papain-like Protease.
Int J Mol Sci, 24, 2023
|
|
7VP2
 
 | | Structure of a transcription factor and DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*GP*TP*GP*GP*TP*CP*CP*CP*CP*AP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*TP*GP*GP*GP*GP*AP*CP*CP*AP*CP*A)-3'), Transcription factor TCP10 | | Authors: | Zhang, Y, Xu, Y.P, Wang, B, Su, X.D. | | Deposit date: | 2021-10-15 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for DNA recognition by TCP transcription factors
To Be Published
|
|
7Y09
 
 | | Cryo-EM structure of human IgM-Fc in complex with the J chain and the DBL domain of DBLMSP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin J chain, ... | | Authors: | Shen, H, Ji, C, Xiao, J. | | Deposit date: | 2022-06-03 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Plasmodium falciparum has evolved multiple mechanisms to hijack human immunoglobulin M.
Nat Commun, 14, 2023
|
|
7YG2
 
 | | Cryo-EM structure of human IgM-Fc in complex with the J chain and the DBL domain of DBLMSP2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DBLMSP2, ... | | Authors: | Shen, H, Ji, C, Xiao, J. | | Deposit date: | 2022-07-09 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Plasmodium falciparum has evolved multiple mechanisms to hijack human immunoglobulin M.
Nat Commun, 14, 2023
|
|
7Y0J
 
 | |
7Y0H
 
 | |
4V7J
 
 | | Structure of RelE nuclease bound to the 70S ribosome (precleavage state) | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Neubauer, C, Gao, Y.-G, Andersen, K.R, Dunham, C.M, Kelley, A.C, Hentschel, J, Gerdes, K, Ramakrishnan, V, Brodersen, D.E. | | Deposit date: | 2009-11-02 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE.
Cell(Cambridge,Mass.), 139, 2009
|
|
5J4D
 
 | |
6WEO
 
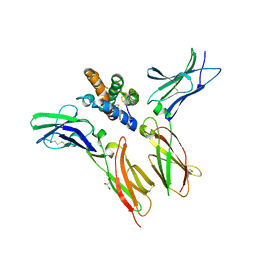 | | IL-22 Signaling Complex with IL-22R1 and IL-10Rbeta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Saxton, R.A, Jude, K.M, Henneberg, L.T, Garcia, K.C. | | Deposit date: | 2020-04-02 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The tissue protective functions of interleukin-22 can be decoupled from pro-inflammatory actions through structure-based design.
Immunity, 54, 2021
|
|
5JC9
 
 | | Structure of the Escherichia coli ribosome with the U1052G mutation in the 16S rRNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, ... | | Authors: | Cocozaki, A, Ferguson, A. | | Deposit date: | 2016-04-14 | | Release date: | 2016-07-06 | | Last modified: | 2016-08-03 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Resistance mutations generate divergent antibiotic susceptibility profiles against translation inhibitors.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8W4U
 
 | | human KCNQ2-CaM in complex with PIP2 and HN37 | | Descriptor: | Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 2, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate, ... | | Authors: | Ma, D, Li, X, Guo, J. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ligand activation mechanisms of human KCNQ2 channel.
Nat Commun, 14, 2023
|
|
4WQ1
 
 | | Complex of 70S ribosome with tRNA-Tyr and mRNA with C-A mismatch in the first position in the A-site. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2014-10-21 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural insights into the translational infidelity mechanism.
Nat Commun, 6, 2015
|
|
4WRO
 
 | | Complex of 70S ribosome with tRNA-Phe and mRNA with C-A mismatch in the second position in the A-site | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2014-10-24 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural insights into the translational infidelity mechanism.
Nat Commun, 6, 2015
|
|
4WSD
 
 | | Complex of 70S ribosome with tRNA-Phe and mRNA with C-A mismatch in the second position in the A-site and with antibiotic paromomycin. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2014-10-27 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural insights into the translational infidelity mechanism.
Nat Commun, 6, 2015
|
|
4WSM
 
 | | Complex of 70S ribosome with tRNA-Leu and mRNA with G-U mismatch in the first position in the A- and P-sites | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2014-10-28 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural insights into the translational infidelity mechanism.
Nat Commun, 6, 2015
|
|
4WQR
 
 | | Complex of 70S ribosome with tRNA-Phe and mRNA with C-A mismatch in the first position in the A-site. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2014-10-22 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural insights into the translational infidelity mechanism.
Nat Commun, 6, 2015
|
|
4WT1
 
 | | Complex of 70S ribosome with tRNA-Phe and mRNA with A-A mismatch in the second position in the A-site | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2014-10-29 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural insights into the translational infidelity mechanism.
Nat Commun, 6, 2015
|
|
8YD8
 
 | |
4V5K
 
 | | Structure of cytotoxic domain of colicin E3 bound to the 70S ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ng, C.L, Lang, K, Meenan, N.A.G, Sharma, A, Kelley, A.C, Kleanthous, C, Ramakrishnan, V. | | Deposit date: | 2010-05-29 | | Release date: | 2014-07-09 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis for 16S Ribosomal RNA Cleavage by the Cytotoxic Domain of Colicin E3.
Nat.Struct.Mol.Biol., 17, 2010
|
|
