6FQU
 
 | | Crystal structure of CREBBP bromodomain complexd with DR09 | | Descriptor: | 1-[3-[3-[3,3-bis(fluoranyl)piperidin-1-yl]phenyl]-4-ethoxy-phenyl]ethanone, CREB-binding protein | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2018-02-14 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
6FR0
 
 | | Crystal structure of CREBBP bromodomain complexd with PB08 | | Descriptor: | CREB-binding protein, ~{N}-[3-(5-ethanoyl-2-ethoxy-phenyl)-5-(2-ethyl-5-methyl-3-oxidanylidene-1,2-oxazol-4-yl)phenyl]furan-2-carboxamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2018-02-15 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
5OWK
 
 | |
3FCS
 
 | | Structure of complete ectodomain of integrin aIIBb3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Zhu, J, Luo, B.-H, Xiao, T, Zhang, C, Nishida, N, Springer, T.A. | | Deposit date: | 2008-11-22 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces.
Mol.Cell, 32, 2008
|
|
3RJR
 
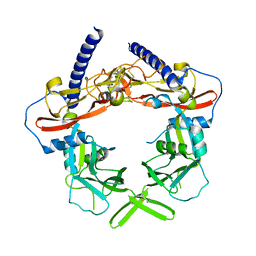 | | Crystal Structure of pro-TGF beta 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Transforming growth factor beta-1 | | Authors: | Zhu, J.H, Shi, M.L, Springer, T.A. | | Deposit date: | 2011-04-15 | | Release date: | 2011-06-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Latent TGF-Beta structure and activation
Nature, 474, 2011
|
|
5OWW
 
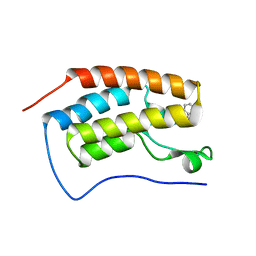 | | Crystal structure of human BRD4(1) bromodomain in complex with UT22B | | Descriptor: | Bromodomain-containing protein 4, ~{N}-(3-methylbenzotriazol-5-yl)-1-(phenylmethyl)imidazole-2-carboxamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2017-09-04 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Chemical Space Expansion of Bromodomain Ligands Guided by in Silico Virtual Couplings (AutoCouple).
Acs Cent.Sci., 4, 2018
|
|
7B55
 
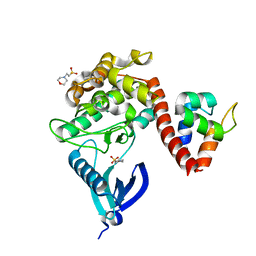 | | Crystal structure of CaMKII-actinin complex bound to MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Alpha-actinin-2, Calcium/calmodulin-dependent protein kinase type II subunit alpha | | Authors: | Zhu, J, Gold, M. | | Deposit date: | 2020-12-03 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of CaMKII-actinin complex bound to MES
To Be Published
|
|
7B56
 
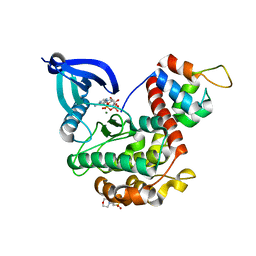 | | Crystal structure of CaMKII-actinin complex bound to AMPPNP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Alpha-actinin-2, Calcium/calmodulin-dependent protein kinase type II subunit alpha, ... | | Authors: | Zhu, J, Gold, M. | | Deposit date: | 2020-12-03 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of CaMKII-actinin complex bound to MES
To Be Published
|
|
7B57
 
 | | Crystal structure of CaMKII-actinin complex bound to ADP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, Alpha-actinin-2, ... | | Authors: | Zhu, J, Gold, M. | | Deposit date: | 2020-12-03 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of CaMKII-actinin complex bound to ADP
To Be Published
|
|
5COD
 
 | | Bovine heart complex I membrane domain | | Descriptor: | NADH-ubiquinone oxidoreductase chain 4, NADH-ubiquinone oxidoreductase chain 5, SDAP, ... | | Authors: | Zhu, J, Hirst, J, King, M.S, Yu, M, Leslie, A.G.W, Klipcan, L. | | Deposit date: | 2015-07-20 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (6.74 Å) | | Cite: | Structure of subcomplex I beta of mammalian respiratory complex I leads to new supernumerary subunit assignments.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5D7X
 
 | |
3SWE
 
 | | Haemophilus influenzae MurA in complex with UDP-N-acetylmuramic acid and covalent adduct of PEP with Cys117 | | Descriptor: | (2R)-2-{[(2R,3R,4R,5S,6R)-3-(acetylamino)-2-{[(S)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-4-yl]oxy}propanoic acid, GLYCEROL, SULFATE ION, ... | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2011-07-13 | | Release date: | 2012-03-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional Consequence of Covalent Reaction of Phosphoenolpyruvate with UDP-N-acetylglucosamine 1-Carboxyvinyltransferase (MurA).
J.Biol.Chem., 287, 2012
|
|
3SWD
 
 | | E. coli MurA in complex with UDP-N-acetylmuramic acid and covalent adduct of PEP with Cys115 | | Descriptor: | (2R)-2-{[(2R,3R,4R,5S,6R)-3-(acetylamino)-2-{[(S)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-4-yl]oxy}propanoic acid, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2011-07-13 | | Release date: | 2012-03-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Functional Consequence of Covalent Reaction of Phosphoenolpyruvate with UDP-N-acetylglucosamine 1-Carboxyvinyltransferase (MurA).
J.Biol.Chem., 287, 2012
|
|
3RO3
 
 | | crystal structure of LGN/mInscuteable complex | | Descriptor: | CHLORIDE ION, ETHANOL, G-protein-signaling modulator 2, ... | | Authors: | Zhu, J, Wen, W, Shang, Y, Wei, Z, Pan, Z, Wang, W, Zhang, M. | | Deposit date: | 2011-04-25 | | Release date: | 2012-03-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | LGN/mInsc and LGN/NuMA complex structures suggest distinct functions in asymmetric cell division for the Par3/mInsc/LGN and G[alpha]i/LGN/NuMA pathways
Mol.Cell, 43, 2011
|
|
6FQT
 
 | | Crystal structure of CREBBP bromodomain complexd with DR46 | | Descriptor: | CREB-binding protein, ~{N}-[3-(5-ethanoyl-2-ethoxy-phenyl)-5-(1-methylpyrazol-3-yl)phenyl]furan-2-carboxamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2018-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of CREBBP bromodomain complexd with DR46
To Be Published
|
|
5NLK
 
 | | Crystal structure of CREBBP bromodomain complexd with US13A | | Descriptor: | CREB-binding protein, ~{N}-[3-acetamido-5-[(5-ethanoyl-2-ethoxy-phenyl)carbamoyl]phenyl]furan-2-carboxamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2017-04-04 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Chemical Space Expansion of Bromodomain Ligands Guided by in Silico Virtual Couplings (AutoCouple).
ACS Cent Sci, 4, 2018
|
|
6TS3
 
 | |
5OWB
 
 | |
5OVB
 
 | | Crystal structure of human BRD4(1) bromodomain in complex with DR46 | | Descriptor: | Bromodomain-containing protein 4, ~{N}-[3-(5-ethanoyl-2-ethoxy-phenyl)-5-(1-methylpyrazol-3-yl)phenyl]furan-2-carboxamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2017-08-28 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Chemical Space Expansion of Bromodomain Ligands Guided by in Silico Virtual Couplings (AutoCouple).
Acs Cent.Sci., 4, 2018
|
|
5OWE
 
 | |
5OWM
 
 | |
5C85
 
 | | Crystal structure of the human BRPF1 bromodomain in complex with SEED1 | | Descriptor: | 6-bromo-3,4-dihydroquinoxalin-2(1H)-one, NITRATE ION, Peregrin | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2015-06-25 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Twenty Crystal Structures of Bromodomain and PHD Finger Containing Protein 1 (BRPF1)/Ligand Complexes Reveal Conserved Binding Motifs and Rare Interactions.
J.Med.Chem., 59, 2016
|
|
5C7N
 
 | |
5C87
 
 | |
5EWD
 
 | |
