4F83
 
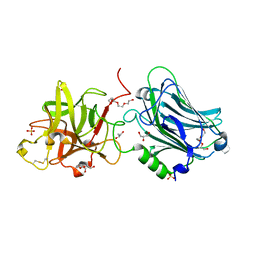 | | Crystal structure of the receptor binding domain of botulinum neurotoxin mosaic serotype C/D with a tetraethylene glycol molecule bound on the Hcn sub-domain and a sulfate ion at the putative active site | | Descriptor: | GLYCEROL, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Zhang, Y, Buchko, G.W, Gardberg, A, Edwards, T.E, Sankaran, B, Robinson, H, Varnum, S.M, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2012-05-16 | | Release date: | 2012-06-20 | | Last modified: | 2013-06-12 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the functional role of the Hcn sub-domain of the receptor-binding domain of the botulinum neurotoxin mosaic serotype C/D.
Biochimie, 95, 2013
|
|
7EN7
 
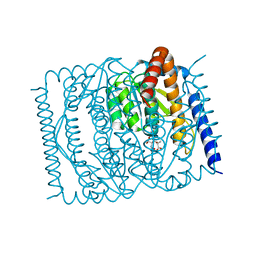 | | The crystal structure of Escherichia coli MurR in complex with N-acetylmuramic-acid-6-phosphate | | Descriptor: | (2R)-2-[(2R,3R,4R,5S,6R)-3-acetamido-2,5-bis(oxidanyl)-6-(phosphonooxymethyl)oxan-4-yl]oxypropanoic acid, HTH-type transcriptional regulator MurR | | Authors: | Zhang, Y, Chen, W, Ji, Q. | | Deposit date: | 2021-04-16 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Molecular basis for cell-wall recycling regulation by transcriptional repressor MurR in Escherichia coli.
Nucleic Acids Res., 50, 2022
|
|
7EN5
 
 | | The crystal structure of Escherichia coli MurR in complex with N-acetylglucosamine-6-phosphate | | Descriptor: | 2-METHOXYETHANOL, 2-acetamido-2-deoxy-6-O-phosphono-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Zhang, Y, Chen, W, Ji, Q. | | Deposit date: | 2021-04-16 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Molecular basis for cell-wall recycling regulation by transcriptional repressor MurR in Escherichia coli.
Nucleic Acids Res., 50, 2022
|
|
4GKU
 
 | |
6BGN
 
 | | Crystal Structure of 4-Oxalocrotonate Tautomerase After Incubation with 5-Fluoro-2-hydroxy-2,4-pentadienoate | | Descriptor: | 2-hydroxymuconate tautomerase, 5-fluoranyl-2-oxidanylidene-pentanoic acid, GLYCEROL, ... | | Authors: | Zhang, Y, Li, W, Stack, T. | | Deposit date: | 2017-10-29 | | Release date: | 2018-02-21 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Inactivation of 4-Oxalocrotonate Tautomerase by 5-Halo-2-hydroxy-2,4-pentadienoates.
Biochemistry, 57, 2018
|
|
3PQE
 
 | |
3PQF
 
 | |
3PME
 
 | | Crystal structure of the receptor binding domain of botulinum neurotoxin C/D mosaic serotype | | Descriptor: | GLYCEROL, SULFATE ION, Type C neurotoxin | | Authors: | Zhang, Y, Buchko, G.W, Qin, L, Robinson, H, Varnum, S.M, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2010-11-16 | | Release date: | 2010-12-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of the receptor binding domain of the botulinum C-D mosaic neurotoxin reveals potential roles of lysines 1118 and 1136 in membrane interactions.
Biochem.Biophys.Res.Commun., 404, 2011
|
|
3PQD
 
 | |
5XS9
 
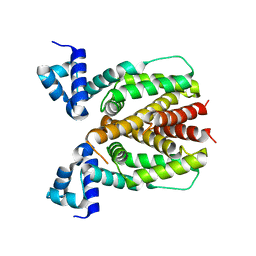 | | Crystal structure of Mycobacterium smegmatis BioQ | | Descriptor: | TetR family transcriptional regulator | | Authors: | Zhang, Y, Ji, Q, Feng, Y. | | Deposit date: | 2017-06-13 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of BioQ suggests a distinct regulatory mechanism for biotin, a nutritional virulence factor in Mycobacterium
To Be Published
|
|
5X87
 
 | |
7EN6
 
 | | The crystal structure of Escherichia coli MurR in apo form | | Descriptor: | HTH-type transcriptional regulator MurR, PHOSPHATE ION | | Authors: | Zhang, Y, Chen, W, Ji, Q. | | Deposit date: | 2021-04-16 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.276 Å) | | Cite: | Molecular basis for cell-wall recycling regulation by transcriptional repressor MurR in Escherichia coli.
Nucleic Acids Res., 50, 2022
|
|
7WFY
 
 | |
8IHT
 
 | | Rpd3S bound to the nucleosome | | Descriptor: | CALCIUM ION, Chromatin modification-related protein EAF3, DNA (164-MER), ... | | Authors: | Zhang, Y, Gang, C. | | Deposit date: | 2023-02-23 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Structural basis for nucleosome binding and catalysis by the yeast Rpd3S/HDAC holoenzyme.
Cell Res., 33, 2023
|
|
8IHM
 
 | | Eaf3 CHD domain bound to the nucleosome | | Descriptor: | Chromatin modification-related protein EAF3, DNA (164-MER), DNA (165-MER), ... | | Authors: | Zhang, Y, Gang, C. | | Deposit date: | 2023-02-23 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural basis for nucleosome binding and catalysis by the yeast Rpd3S/HDAC holoenzyme.
Cell Res., 33, 2023
|
|
8IHN
 
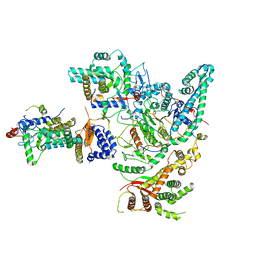 | | Cryo-EM structure of the Rpd3S core complex | | Descriptor: | CALCIUM ION, Chromatin modification-related protein EAF3, Histone H3, ... | | Authors: | Zhang, Y, Gang, C. | | Deposit date: | 2023-02-23 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structural basis for nucleosome binding and catalysis by the yeast Rpd3S/HDAC holoenzyme.
Cell Res., 33, 2023
|
|
7YIS
 
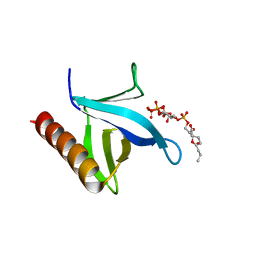 | | Crystal structure of N-terminal PH domain of ARAP3 protein in complex with inositol 1,3,4,5-tetrakisphosphate | | Descriptor: | (2R)-3-{[(S)-{[(2S,3R,5S,6S)-2,6-DIHYDROXY-3,4,5-TRIS(PHOSPHONOOXY)CYCLOHEXYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-(1-HYDROXY BUTOXY)PROPYL BUTYRATE, Arf-GAP with Rho-GAP domain, ANK repeat and PH domain-containing protein 3 | | Authors: | Zhang, Y.J, Liu, Y.R, Wu, B. | | Deposit date: | 2022-07-18 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Insights Uncover the Specific Phosphoinositide Recognition by the PH1 Domain of Arap3.
Int J Mol Sci, 24, 2023
|
|
7YIR
 
 | |
7DRA
 
 | |
7DR8
 
 | | Crystal structure of MERS-CoV 3CL protease in spacegroup P212121 | | Descriptor: | 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2020-12-26 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.338149 Å) | | Cite: | Crystal structure of MERS-CoV 3CL protease in spacegroup P212121
To Be Published
|
|
7ESD
 
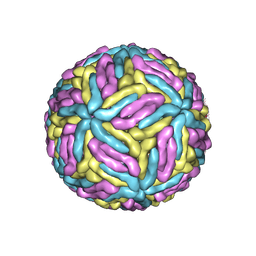 | | Mature Donggang virus | | Descriptor: | Genome polyprotein | | Authors: | Zhang, Y, Liang, D. | | Deposit date: | 2021-05-10 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Replication is the key barrier during the dual-host adaptation of mosquito-borne flaviviruses.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XQV
 
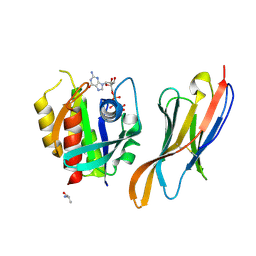 | | The complex of nanobody Rh57 binding to GTP-bound RhoA active form | | Descriptor: | ALANINE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Zhang, Y.R, Liu, R, Ding, Y. | | Deposit date: | 2022-05-09 | | Release date: | 2022-07-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural insights into the binding of nanobody Rh57 to active RhoA-GTP.
Biochem.Biophys.Res.Commun., 616, 2022
|
|
7XP0
 
 | |
7XP1
 
 | | Crystal structure of PmiR from Pseudomonas aeruginosa | | Descriptor: | ALPHA-METHYLISOCITRIC ACID, GLYCEROL, Probable transcriptional regulator, ... | | Authors: | Zhang, Y.X, Liang, H.H, Gan, J.H. | | Deposit date: | 2022-05-02 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | PmiR senses 2-methylisocitrate levels to regulate bacterial virulence in Pseudomonas aeruginosa.
Sci Adv, 8, 2022
|
|
6LN5
 
 | | CryoEM structure of SERCA2b T1032stop in E1-2Ca2+-AMPPCP (class1) | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Zhang, Y, Tsutsumi, A, Watanabe, S, Inaba, K. | | Deposit date: | 2019-12-28 | | Release date: | 2020-08-26 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of SERCA2b reveal the mechanism of regulation by the luminal extension tail.
Sci Adv, 6, 2020
|
|
