1XZ3
 
 | | Complex of apoferritin with isoflurane | | Descriptor: | 1-CHLORO-2,2,2-TRIFLUOROETHYL DIFLUOROMETHYL ETHER, CADMIUM ION, Ferritin light chain | | Authors: | Liu, R, Loll, P.J, Eckenhoff, R.G. | | Deposit date: | 2004-11-11 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for high-affinity volatile anesthetic binding in a natural 4-helix bundle protein.
Faseb J., 19, 2005
|
|
1XZ1
 
 | | Complex of halothane with apoferritin | | Descriptor: | 2-BROMO-2-CHLORO-1,1,1-TRIFLUOROETHANE, CADMIUM ION, Ferritin light chain | | Authors: | Liu, R, Loll, P.J, Eckenhoff, R.G. | | Deposit date: | 2004-11-11 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for high-affinity volatile anesthetic binding in a natural 4-helix bundle protein.
Faseb J., 19, 2005
|
|
3PZ0
 
 | | The crystal structure of AaLeuRS-CP1 | | Descriptor: | CALCIUM ION, Leucyl-tRNA synthetase subunit alpha | | Authors: | Liu, R.J, Wang, E.D. | | Deposit date: | 2010-12-13 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Peripheral insertion modulates the editing activity of the isolated CP1 domain of leucyl-tRNA synthetase
Biochem.J., 440, 2011
|
|
4XBO
 
 | | Crystal structure of full length E.coli TrmJ in complex with SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, tRNA (cytidine/uridine-2'-O-)-methyltransferase TrmJ | | Authors: | Liu, R.J, Long, T, Zhou, M, Wang, E.D. | | Deposit date: | 2014-12-17 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | tRNA recognition by a bacterial tRNA Xm32 modification enzyme from the SPOUT methyltransferase superfamily
Nucleic Acids Res., 43, 2015
|
|
7BZJ
 
 | | The Discovery of Benzhydrol-Oxaborole Hybrid Derivatives as Leucyl-tRNA Synthetase Inhibitors | | Descriptor: | Leucine--tRNA ligase, [(1~{R},5~{R},6~{S},8~{R})-8-(6-aminopurin-9-yl)-4'-[(~{R})-oxidanyl-[4-(2-oxidanylidenepropylsulfanyl)phenyl]methyl]spiro[2,4,7-trioxa-3-boranuidabicyclo[3.3.0]octane-3,7'-7-boranuidabicyclo[4.3.0]nona-1(6),2,4-triene]-6-yl]methoxy-tris(oxidanyl)phosphanium | | Authors: | Liu, R.J, Li, H, Wang, E.D, Zhou, H. | | Deposit date: | 2020-04-28 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of benzhydrol-oxaborole derivatives as Streptococcus pneumoniae leucyl-tRNA synthetase inhibitors.
Bioorg.Med.Chem., 29, 2021
|
|
3PZ5
 
 | | The crystal structure of AaLeuRS-CP1-D20 | | Descriptor: | Leucyl-tRNA synthetase subunit alpha | | Authors: | Liu, R.J, Wang, E.D. | | Deposit date: | 2010-12-14 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Peripheral insertion modulates the editing activity of the isolated CP1 domain of leucyl-tRNA synthetase
Biochem.J., 440, 2011
|
|
4JAK
 
 | |
4JAL
 
 | | Crystal structure of tRNA (Um34/Cm34) methyltransferase TrmL from Escherichia coli with SAH | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Liu, R.J, Zhou, M, Wang, E.D. | | Deposit date: | 2013-02-18 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The tRNA recognition mechanism of the minimalist SPOUT methyltransferase, TrmL
Nucleic Acids Res., 41, 2013
|
|
1QS9
 
 | |
1QTD
 
 | |
1QS5
 
 | |
1QSB
 
 | |
1QTH
 
 | |
1QTC
 
 | |
1QTB
 
 | |
7VSQ
 
 | |
3U90
 
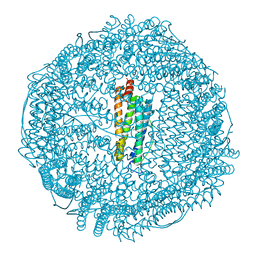 | | apoferritin: complex with SDS | | Descriptor: | CADMIUM ION, DODECYL SULFATE, Ferritin light chain | | Authors: | Liu, R, Bu, W, Xi, J, Mortazavi, S.R, Cheung-Lau, J.C, Dmochowski, I.J, Loll, P.J. | | Deposit date: | 2011-10-17 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Beyond the detergent effect: a binding site for sodium dodecyl sulfate (SDS) in mammalian apoferritin.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3NDP
 
 | | Crystal structure of human AK4(L171P) | | Descriptor: | Adenylate kinase isoenzyme 4, SULFATE ION | | Authors: | Liu, R, Wang, Y, Wei, Z, Gong, W. | | Deposit date: | 2010-06-07 | | Release date: | 2010-06-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human adenylate kinase 4 (L171P) suggests the role of hinge region in protein domain motion
Biochem.Biophys.Res.Commun., 379, 2009
|
|
3PZ6
 
 | | The crystal structure of GlLeuRS-CP1 | | Descriptor: | Leucyl-tRNA synthetase | | Authors: | Liu, R.J, Du, D.H, Wang, E.D. | | Deposit date: | 2010-12-14 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Peripheral insertion modulates the editing activity of the isolated CP1 domain of leucyl-tRNA synthetase
Biochem.J., 440, 2011
|
|
5WWR
 
 | | Crystal structure of human NSun6/tRNA/SFG | | Descriptor: | Putative methyltransferase NSUN6, SINEFUNGIN, tRNA | | Authors: | Liu, R.J, Long, T, Wang, E.D. | | Deposit date: | 2017-01-04 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.096 Å) | | Cite: | Structural basis for substrate binding and catalytic mechanism of a human RNA:m5C methyltransferase NSun6
Nucleic Acids Res., 45, 2017
|
|
5WWT
 
 | | Crystal structure of human NSun6/tRNA | | Descriptor: | Putative methyltransferase NSUN6, tRNA | | Authors: | Liu, R.J, Long, T, Wang, E.D. | | Deposit date: | 2017-01-04 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.197 Å) | | Cite: | Structural basis for substrate binding and catalytic mechanism of a human RNA:m5C methyltransferase NSun6
Nucleic Acids Res., 45, 2017
|
|
5WWS
 
 | | Crystal structure of human NSun6/tRNA/SAM | | Descriptor: | Putative methyltransferase NSUN6, S-ADENOSYLMETHIONINE, tRNA | | Authors: | Liu, R.J, Long, T, Wang, E.D. | | Deposit date: | 2017-01-04 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.247 Å) | | Cite: | Structural basis for substrate binding and catalytic mechanism of a human RNA:m5C methyltransferase NSun6
Nucleic Acids Res., 45, 2017
|
|
5WWQ
 
 | | Crystal structure of human NSun6 | | Descriptor: | Putative methyltransferase NSUN6 | | Authors: | Liu, R.J, Long, T, Wang, E.D. | | Deposit date: | 2017-01-04 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.815 Å) | | Cite: | Structural basis for substrate binding and catalytic mechanism of a human RNA:m5C methyltransferase NSun6
Nucleic Acids Res., 45, 2017
|
|
5YXI
 
 | | Designed protein dRafX6 | | Descriptor: | Designed protein dRafX6 | | Authors: | Liu, R. | | Deposit date: | 2017-12-05 | | Release date: | 2018-12-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | De novo sequence redesign of a functional Ras-binding domain globally inverted the surface charge distribution and led to extreme thermostability.
Biotechnol.Bioeng., 118, 2021
|
|
6LQE
 
 | |
