7YWX
 
 | | Structure of the human CCAN CENP-A alpha-satellite complex | | Descriptor: | Centromere protein C, Centromere protein H, Centromere protein I, ... | | Authors: | Yatskevich, S, Muir, K.W, Bellini, D, Zhang, Z, Yang, J, Tischer, T, Predin, M, Dendooven, T, McLaughlin, S.H, Barford, D. | | Deposit date: | 2022-02-14 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structure of the human inner kinetochore bound to a centromeric CENP-A nucleosome.
Science, 376, 2022
|
|
2ASQ
 
 | | Solution Structure of SUMO-1 in Complex with a SUMO-binding Motif (SBM) | | Descriptor: | Protein inhibitor of activated STAT2, Small ubiquitin-related modifier 1 | | Authors: | Song, J, Zhang, Z, Hu, W, Chen, Y. | | Deposit date: | 2005-08-23 | | Release date: | 2005-10-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Small Ubiquitin-like Modifier (SUMO) Recognition of a SUMO Binding Motif: A reversal of the bound orientation
J.Biol.Chem., 280, 2005
|
|
8A61
 
 | | S. cerevisiae apo phosphorylated APC/C | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 11, Anaphase-promoting complex subunit 2, ... | | Authors: | Barford, D, Fernandez-Vazquez, E, Zhang, Z, Yang, J. | | Deposit date: | 2022-06-16 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Cryo-EM structure of the S. cerevisiae apo phosphorylated APC/C
To Be Published
|
|
8A3T
 
 | | S. cerevisiae APC/C-Cdh1 complex | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 11, Anaphase-promoting complex subunit 2, ... | | Authors: | Barford, D, Vazquez-Fernandez, E, Zhang, Z, Yang, J. | | Deposit date: | 2022-06-09 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of the S. cerevisiae APC/C-Cdh1 complex
To Be Published
|
|
6GYU
 
 | | Cryo-EM structure of the CBF3-msk complex of the budding yeast kinetochore | | Descriptor: | Centromere DNA-binding protein complex CBF3 subunit A, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | Authors: | Yan, K, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | Deposit date: | 2018-07-02 | | Release date: | 2018-12-05 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Architecture of the CBF3-centromere complex of the budding yeast kinetochore.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6TGB
 
 | | CryoEM structure of the binary DOCK2-ELMO1 complex | | Descriptor: | Dedicator of cytokinesis protein 2, Engulfment and cell motility protein 1 | | Authors: | Chang, L, Yang, J, Chang, J.H, Zhang, Z, Boland, A, McLaughlin, S.H, Abu-Thuraia, A, Killoran, R.C, Smith, M.J, Cote, J.F, Barford, D. | | Deposit date: | 2019-11-15 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structure of the DOCK2-ELMO1 complex provides insights into regulation of the auto-inhibited state.
Nat Commun, 11, 2020
|
|
8A5Y
 
 | | S. cerevisiae apo unphosphorylated APC/C. | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 11, Anaphase-promoting complex subunit 2, ... | | Authors: | Barford, D, Fernandez-Vazquez, E, Zhang, Z, Yang, J. | | Deposit date: | 2022-06-16 | | Release date: | 2022-08-31 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Cryo-EM structure of the S. cerevisiae APC/C-Cdh1 complex and comparison to apo unphosphorylated and phosphorylated states
To Be Published
|
|
6GYP
 
 | | Cryo-EM structure of the CBF3-core-Ndc10-DBD complex of the budding yeast kinetochore | | Descriptor: | ARGININE, ASPARAGINE, Centromere DNA-binding protein complex CBF3 subunit A, ... | | Authors: | Yan, K, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | Deposit date: | 2018-07-01 | | Release date: | 2018-12-05 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Architecture of the CBF3-centromere complex of the budding yeast kinetochore.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6GYS
 
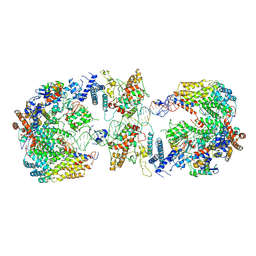 | | Cryo-EM structure of the CBF3-CEN3 complex of the budding yeast kinetochore | | Descriptor: | Centromere DNA-binding protein complex CBF3 subunit A, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | Authors: | Yan, K, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | Deposit date: | 2018-07-01 | | Release date: | 2018-12-05 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Architecture of the CBF3-centromere complex of the budding yeast kinetochore.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6TGC
 
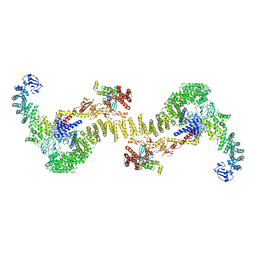 | | CryoEM structure of the ternary DOCK2-ELMO1-RAC1 complex. | | Descriptor: | Dedicator of cytokinesis protein 2, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Chang, L, Yang, J, Chang, J.H, Zhang, Z, Boland, A, McLaughlin, S.H, Abu-Thuraia, A, Killoran, R.C, Smith, M.J, Cote, J.F, Barford, D. | | Deposit date: | 2019-11-15 | | Release date: | 2020-07-29 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the DOCK2-ELMO1 complex provides insights into regulation of the auto-inhibited state.
Nat Commun, 11, 2020
|
|
2E9W
 
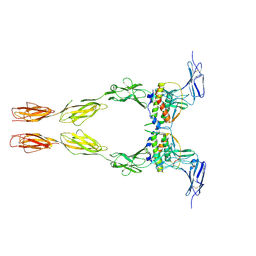 | | Crystal structure of the extracellular domain of Kit in complex with stem cell factor (SCF) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Kit ligand, Mast/stem cell growth factor receptor | | Authors: | Yuzawa, S, Opatowsky, Y, Zhang, Z, Mandiyan, V, Lax, I, Schlessinger, J. | | Deposit date: | 2007-01-27 | | Release date: | 2007-08-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Basis for Activation of the Receptor Tyrosine Kinase KIT by Stem Cell Factor
Cell(Cambridge,Mass.), 130, 2007
|
|
2EC8
 
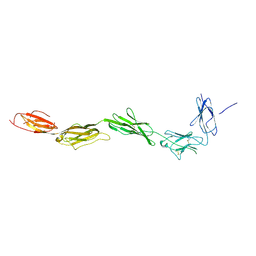 | | Crystal structure of the exctracellular domain of the receptor tyrosine kinase, Kit | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Mast/stem cell growth factor receptor | | Authors: | Yuzawa, S, Opatowsky, Y, Zhang, Z, Mandiyan, V, Lax, I, Schlessinger, J. | | Deposit date: | 2007-02-11 | | Release date: | 2007-08-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Activation of the Receptor Tyrosine Kinase KIT by Stem Cell Factor
Cell(Cambridge,Mass.), 130, 2007
|
|
2F7L
 
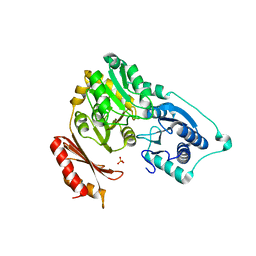 | | Crystal structure of Sulfolobus tokodaii phosphomannomutase/phosphoglucomutase | | Descriptor: | 455aa long hypothetical phospho-sugar mutase, SULFATE ION | | Authors: | Kawamura, T, Sakai, N, Akutsu, J, Zhang, Z, Watanabe, N, Kawarabayashi, Y, Tanaka, I. | | Deposit date: | 2005-12-01 | | Release date: | 2006-12-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Sulfolobus tokodaii phosphomannomutase/phosphoglucomutase
To be Published
|
|
2FOT
 
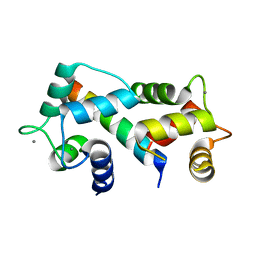 | | Crystal structure of the complex between calmodulin and alphaII-spectrin | | Descriptor: | CALCIUM ION, Calmodulin, alpha-II spectrin Spectrin | | Authors: | Simonovic, M, Zhang, Z, Cianci, C.D, Steitz, T.A, Morrow, J.S. | | Deposit date: | 2006-01-13 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the calmodulin alphaII-spectrin complex provides insight into the regulation of cell plasticity.
J.Biol.Chem., 281, 2006
|
|
5MOF
 
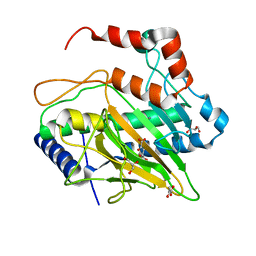 | | Ethylene Forming Enzyme from Pseudomonas syringae pv. phaseolicola - I222 crystal form in complex with manganese and 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, CHLORIDE ION, ... | | Authors: | McDonough, M.A, Zhang, Z, Schofield, C.J. | | Deposit date: | 2016-12-14 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and stereoelectronic insights into oxygenase-catalyzed formation of ethylene from 2-oxoglutarate.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4BH6
 
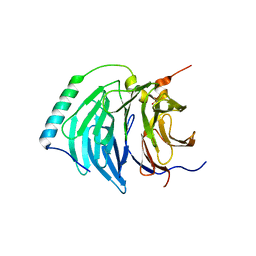 | | Insights into degron recognition by APC coactivators from the structure of an Acm1-Cdh1 complex | | Descriptor: | APC/C ACTIVATOR PROTEIN CDH1, APC/C-CDH1 MODULATOR 1 | | Authors: | He, J, Chao, W.C.H, Zhang, Z, Yang, J, Cronin, N, Barford, D. | | Deposit date: | 2013-03-29 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insights Into Degron Recognition by Apc/C Coactivators from the Structure of an Acm1-Cdh1 Complex.
Mol.Cell, 50, 2013
|
|
5MZ6
 
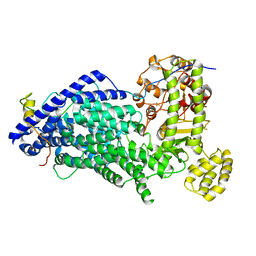 | | Cryo-EM structure of a Separase-Securin complex from Caenorhabditis elegans at 3.8 A resolution | | Descriptor: | Interactor of FizzY protein, SEParase | | Authors: | Boland, A, Martin, T.G, Zhang, Z, Yang, J, Bai, X.C, Chang, L, Scheres, S.H.W, Barford, D. | | Deposit date: | 2017-01-31 | | Release date: | 2017-03-08 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of a metazoan separase-securin complex at near-atomic resolution.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5N7Z
 
 | |
1JOT
 
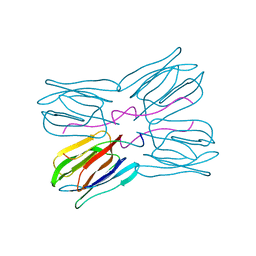 | | STRUCTURE OF THE LECTIN MPA COMPLEXED WITH T-ANTIGEN DISACCHARIDE | | Descriptor: | AGGLUTININ, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose | | Authors: | Lee, X, Thompson, A, Zhang, Z, Hoa, T.-T, Biesterfeldt, J, Ogata, C, Xu, L, Johnston, R.A.Z, Young, N.M. | | Deposit date: | 1997-12-05 | | Release date: | 1998-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the complex of Maclura pomifera agglutinin and the T-antigen disaccharide, Galbeta1,3GalNAc.
J.Biol.Chem., 273, 1998
|
|
1LCY
 
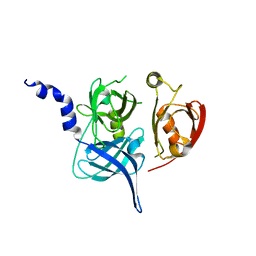 | | Crystal Structure of the Mitochondrial Serine Protease HtrA2 | | Descriptor: | HtrA2 serine protease | | Authors: | Li, W, Srinivasula, S.M, Chai, J, Li, P, Wu, J.W, Zhang, Z, Alnemri, E.S, Shi, Y. | | Deposit date: | 2002-04-07 | | Release date: | 2002-05-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the pro-apoptotic function of mitochondrial serine protease HtrA2/Omi.
Nat.Struct.Biol., 9, 2002
|
|
4E55
 
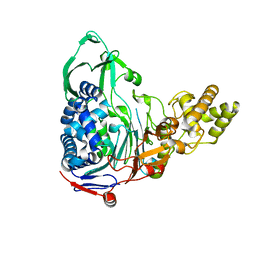 | |
4E57
 
 | |
4E56
 
 | |
4FQ7
 
 | | Crystal structure of the maleate isomerase Iso from Pseudomonas putida S16 | | Descriptor: | Maleate cis-trans isomerase | | Authors: | Lu, Y, Chen, D, Zhang, Z, Li, Q, Wu, G, Xu, P. | | Deposit date: | 2012-06-25 | | Release date: | 2013-07-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and computational studies of the maleate isomerase from Pseudomonas putida S16 reveal a breathing motion wrapping the substrate inside.
Mol.Microbiol., 87, 2013
|
|
4FQ5
 
 | | Crystal structure of the maleate isomerase Iso(C200A) from Pseudomonas putida S16 with maleate | | Descriptor: | MALEIC ACID, Maleate cis-trans isomerase | | Authors: | Lu, Y, Chen, D, Zhang, Z, Li, Q, Wu, G, Xu, P. | | Deposit date: | 2012-06-25 | | Release date: | 2013-07-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and computational studies of the maleate isomerase from Pseudomonas putida S16 reveal a breathing motion wrapping the substrate inside.
Mol.Microbiol., 87, 2013
|
|
