4LV9
 
 | | Fragment-based Identification of Amides Derived From trans-2-(Pyridin-3-yl)cyclopropanecarboxylic Acid as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 1,2-ETHANEDIOL, 7-chloro-3-methyl-2H-1,2,4-benzothiadiazine 1,1-dioxide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Giannetti, A.M, Zheng, X, Skelton, N, Wang, W, Bravo, B, Feng, Y, Gunzner-Toste, J, Ho, Y, Hua, R, Wang, C, Zhao, Q, Liederer, B.M, Liu, Y, O'Brien, T, Oeh, J, Sampath, D, Shen, Y, Wang, L, Wu, H, Xiao, Y, Yuen, P, Zak, M, Zhao, G, Dragovich, P.S. | | Deposit date: | 2013-07-26 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.807 Å) | | Cite: | Identification of amides derived from 1H-pyrazolo[3,4-b]pyridine-5-carboxylic acid as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4LVF
 
 | | Fragment-based Identification of Amides Derived From trans-2-(Pyridin-3-yl)cyclopropanecarboxylic Acid as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | (1S,2S)-2-phenyl-N-(pyridin-4-yl)cyclopropanecarboxamide, 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Giannetti, A.M, Zheng, X, Skelton, N, Wang, W, Bravo, B, Feng, Y, Gunzner-Toste, J, Ho, Y, Hua, R, Wang, C, Zhao, Q, Liederer, B.M, Liu, Y, O'Brien, T, Oeh, J, Sampath, D, Shen, Y, Wang, L, Wu, H, Xiao, Y, Yuen, P, Zak, M, Zhao, G, Dragovich, P.S. | | Deposit date: | 2013-07-26 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of amides derived from 1H-pyrazolo[3,4-b]pyridine-5-carboxylic acid as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2A14
 
 | | Crystal Structure of Human Indolethylamine N-methyltransferase with SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, indolethylamine N-methyltransferase | | Authors: | Dong, A, Wu, H, Zeng, H, Loppnau, P, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-06-17 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Human Indolethylamine
N-methyltransferase in complex with SAH.
To be Published
|
|
4LVA
 
 | | Fragment-based Identification of Amides Derived From trans-2-(Pyridin-3-yl)cyclopropanecarboxylic Acid as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 1,2-ETHANEDIOL, N-(4-{[4-(pyrrolidin-1-yl)piperidin-1-yl]sulfonyl}benzyl)-2H-pyrido[4,3-e][1,2,4]thiadiazin-3-amine 1,1-dioxide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Giannetti, A.M, Zheng, X, Skelton, N, Wang, W, Bravo, B, Feng, Y, Gunzner-Toste, J, Ho, Y, Hua, R, Wang, C, Zhao, Q, Liederer, B.M, Liu, Y, O'Brien, T, Oeh, J, Sampath, D, Shen, Y, Wang, L, Wu, H, Xiao, Y, Yuen, P, Zak, M, Zhao, G, Dragovich, P.S. | | Deposit date: | 2013-07-26 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification of amides derived from 1H-pyrazolo[3,4-b]pyridine-5-carboxylic acid as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2B9E
 
 | | Human NSUN5 protein | | Descriptor: | NOL1/NOP2/Sun domain family, member 5 isoform 2, S-ADENOSYLMETHIONINE | | Authors: | Min, J.R, Wu, H, Zeng, H, Loppnau, P, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-10-11 | | Release date: | 2005-10-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Crystal Structure of Human NSUN5 protein in complex with
S-adenosyl-L-methionine
To be Published
|
|
2OU2
 
 | | Acetyltransferase domain of Human HIV-1 Tat interacting protein, 60kDa, isoform 3 | | Descriptor: | ACETYL COENZYME *A, Histone acetyltransferase HTATIP, ZINC ION | | Authors: | Min, J, Wu, H, Dombrovski, L, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-09 | | Release date: | 2007-02-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of acetyltransferase domain of Human HIV-1 Tat interacting protein in complex with acetylcoenzyme A.
To be Published
|
|
7RAV
 
 | | Cryo-EM structure of the unliganded form of NLR family apoptosis inhibitory protein 5 (NAIP5) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Baculoviral IAP repeat-containing protein 1e | | Authors: | Paidimuddala, B, Cao, J, Xie, Q, Wu, H, Zhang, L. | | Deposit date: | 2021-07-02 | | Release date: | 2023-01-11 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism of NAIP-NLRC4 inflammasome activation revealed by cryo-EM structure of unliganded NAIP5.
Nat.Struct.Mol.Biol., 30, 2023
|
|
2QL1
 
 | | Structural Characterization of a Mutated, ADCC-Enhanced Human Fc Fragment | | Descriptor: | IGHM protein, ZINC ION, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Oganesyan, V, Wu, H, Dall'Acqua, W.F. | | Deposit date: | 2007-07-12 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.534 Å) | | Cite: | Structural characterization of a mutated, ADCC-enhanced human Fc fragment
Mol.Immunol., 45, 2008
|
|
3FX0
 
 | |
2VXK
 
 | | Structural comparison between Aspergillus fumigatus and human GNA1 | | Descriptor: | 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, COENZYME A, GLUCOSAMINE 6-PHOSPHATE ACETYLTRANSFERASE, ... | | Authors: | Hurtado-Guerrero, R, Raimi, O.G, Min, J, Zeng, H, Vallius, L, Shepherd, S, Ibrahim, A.F.M, Wu, H, Plotnikov, A.N, van Aalten, D.M.F. | | Deposit date: | 2008-07-05 | | Release date: | 2008-07-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Kinetic Differences between Human and Aspergillus Fumigatus D-Glucosamine-6- Phosphate N-Acetyltransferase.
Biochem.J., 415, 2008
|
|
3HM5
 
 | | SANT domain of human DNA methyltransferase 1 associated protein 1 | | Descriptor: | CALCIUM ION, DNA methyltransferase 1-associated protein 1, UNKNOWN ATOM OR ION | | Authors: | Dombrovski, L, Tempel, W, Amaya, M.F, Tong, Y, Ni, S, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Park, H, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-05-28 | | Release date: | 2009-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | SANT domain of human DNA methyltransferase 1 associated protein 1
To be Published
|
|
3HNA
 
 | | Crystal structure of catalytic domain of human euchromatic histone methyltransferase 1 in complex with SAH and mono-Methylated H3K9 Peptide | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-9 specific 5, Mono-Methylated H3K9 Peptide, ... | | Authors: | Min, J, Wu, H, Loppnau, P, Wleigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-05-30 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural biology of human H3K9 methyltransferases
Plos One, 5, 2010
|
|
4YVD
 
 | | Crytsal structure of human Pleiotropic Regulator 1 (PRL1) | | Descriptor: | CHLORIDE ION, Pleiotropic regulator 1, SODIUM ION, ... | | Authors: | Dong, A, Zeng, H, Xu, C, Tempel, W, Li, Y, He, H, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-03-19 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crytsal structure of human Pleiotropic Regulator 1 (PRL1).
to be published
|
|
3CKK
 
 | | Crystal structure of human methyltransferase-like protein 1 | | Descriptor: | GLYCEROL, S-ADENOSYLMETHIONINE, tRNA (guanine-N(7)-)-methyltransferase | | Authors: | Dong, A, Zeng, H, Dobrovetsky, E, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Plotnikov, A.N, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-16 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of human methyltransferase-like protein 1.
To be Published
|
|
6CB8
 
 | |
4WUW
 
 | | Crystal Structure of Surfactant Protein-A DED Mutant (E171D/P175E/K203D) Complexed with Inositol | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, CALCIUM ION, Pulmonary surfactant-associated protein A | | Authors: | Rynkiewicz, M.J, Wu, H, Cafarella, T.R, Nikolaidis, N.M, Head, J.F, Seaton, B.A, McCormack, F.X. | | Deposit date: | 2014-11-03 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Differential ligand binding specificities of the pulmonary collectins are determined by the conformational freedom of a surface loop
To be published
|
|
4WR9
 
 | | Crystal Structure of Surfactant Protein-A DED Mutant (E171D/P175E/K203D) | | Descriptor: | CALCIUM ION, Pulmonary surfactant-associated protein A | | Authors: | Rynkiewicz, M.J, Wu, H, Cafarella, T.R, Nikolaidis, N.M, Head, J.F, Seaton, B.A, McCormack, F.X. | | Deposit date: | 2014-10-23 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Differential ligand binding specificities of the pulmonary collectins are determined by the conformational freedom of a surface loop
To be published
|
|
4WUX
 
 | | Crystal Structure of Surfactant Protein-A DED Mutant (E171D/P175E/K203D) Complexed with Mannose | | Descriptor: | CALCIUM ION, Pulmonary surfactant-associated protein A, alpha-D-mannopyranose | | Authors: | Rynkiewicz, M.J, Wu, H, Cafarella, T.R, Nikolaidis, N.M, Head, J.F, Seaton, B.A, McCormack, F.X. | | Deposit date: | 2014-11-03 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Differential ligand binding specificities of the pulmonary collectins are determined by the conformational freedom of a surface loop
To be published
|
|
3EPP
 
 | | Crystal structure of mRNA cap guanine-N7 methyltransferase (RNMT) in complex with sinefungin | | Descriptor: | SINEFUNGIN, mRNA cap guanine-N7 methyltransferase | | Authors: | Amaya, M.F, Zeng, H, Loppnau, P, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Botchkarev, A, Min, J, Plotnikov, A.N, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-09-29 | | Release date: | 2008-10-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal structure of mRNA cap guanine-N7 methyltransferase (RNMT) in complex with sinefungin.
To be Published
|
|
3LLR
 
 | | Crystal structure of the PWWP domain of Human DNA (cytosine-5-)-methyltransferase 3 alpha | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA (cytosine-5)-methyltransferase 3A, SULFATE ION | | Authors: | Qiu, W, Dombrovski, L, Ni, S, Weigelt, J, Boutra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-01-29 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and histone binding ability characterizations of human PWWP domains.
Plos One, 6, 2011
|
|
8JD9
 
 | | Cyro-EM structure of the Na+/H+ antipoter SOS1 from Arabidopsis thaliana,class1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Sodium/hydrogen exchanger 7 | | Authors: | Yang, G.H, Zhang, Y.M, Zhou, J.Q, Jia, Y.T, Xu, X, Fu, P, Wu, H.Y. | | Deposit date: | 2023-05-13 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structural basis for the activity regulation of Salt Overly Sensitive 1 in Arabidopsis salt tolerance.
Nat.Plants, 9, 2023
|
|
8JDA
 
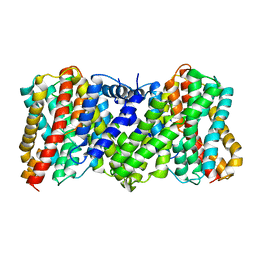 | | Cyro-EM structure of the Na+/H+ antipoter SOS1 from Arabidopsis thaliana,class2 | | Descriptor: | Sodium/hydrogen exchanger 7 | | Authors: | Yang, G.H, Zhang, Y.M, Zhou, J.Q, Jia, Y.T, Xu, X, Fu, P, Wu, H.Y. | | Deposit date: | 2023-05-13 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structural basis for the activity regulation of Salt Overly Sensitive 1 in Arabidopsis salt tolerance.
Nat.Plants, 9, 2023
|
|
3FLG
 
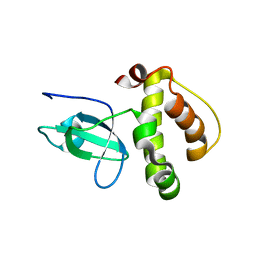 | | The PWWP domain of Human DNA (cytosine-5-)-methyltransferase 3 beta | | Descriptor: | DNA (cytosine-5)-methyltransferase 3B | | Authors: | Amaya, M.F, Zeng, H, MacKenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Botchkarev, A, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-12-18 | | Release date: | 2009-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the PWWP domain of Human DNA (cytosine-5-)-methyltransferase 3 beta
To be Published
|
|
6JON
 
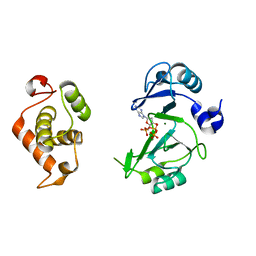 | | Crystal structures of phage NrS-1 N300-dNTPs-Mg2+ complex provide molecular mechanisms for substrate specificity | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, MAGNESIUM ION, Primase | | Authors: | Guo, H.J, Li, M.J, Wu, H, Yu, F, He, J.H. | | Deposit date: | 2019-03-22 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structures of phage NrS-1 N300-dNTPs-Mg2+complex provide molecular mechanisms for substrate specificity.
Biochem.Biophys.Res.Commun., 515, 2019
|
|
6JOP
 
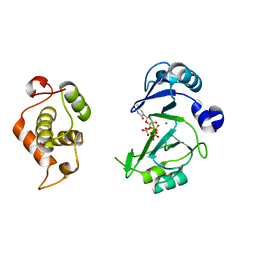 | | Crystal structures of phage NrS-1 N300-dNTPs-Mg2+ complex provide molecular mechanisms for substrate specificity | | Descriptor: | MAGNESIUM ION, Primase, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Guo, H.J, Li, M.J, Wu, H, Yu, F, He, J.H. | | Deposit date: | 2019-03-22 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Crystal structures of phage NrS-1 N300-dNTPs-Mg2+complex provide molecular mechanisms for substrate specificity.
Biochem.Biophys.Res.Commun., 515, 2019
|
|
