2WD1
 
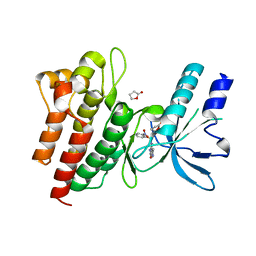 | | Human c-Met Kinase in complex with azaindole inhibitor | | Descriptor: | 1-[(2-NITROPHENYL)SULFONYL]-1H-PYRROLO[3,2-B]PYRIDINE-6-CARBOXAMIDE, GAMMA-BUTYROLACTONE, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | Porter, J, Lumb, S, Franklin, R.J, Gascon-Simorte, J.M, Calmiano, M, Le Riche, K, Lallemand, B, Keyaerts, J, Edwards, H, Maloney, A, Delgado, J, King, L, Foley, A, Lecomte, F, Reuberson, J, Meier, C, Batchelor, M. | | Deposit date: | 2009-03-19 | | Release date: | 2009-04-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of 4-Azaindoles as Novel Inhibitors of C- met Kinase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2W3L
 
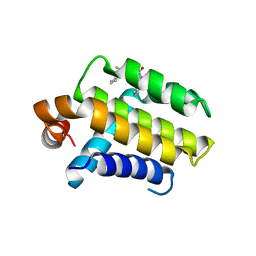 | | Crystal Structure of Chimaeric Bcl2-xL and Phenyl Tetrahydroisoquinoline Amide Complex | | Descriptor: | 1-(2-{[(3S)-3-(aminomethyl)-3,4-dihydroisoquinolin-2(1H)-yl]carbonyl}phenyl)-4-chloro-5-methyl-N,N-diphenyl-1H-pyrazole-3-carboxamide, APOPTOSIS REGULATOR BCL-2 | | Authors: | Porter, J, Payne, A, de Candole, B, Ford, D, Hutchinson, B, Trevitt, G, Turner, J, Edwards, C, Watkins, C, Whitcombe, I, Davis, J, Stubberfield, C, Fisher, M, Lamers, M. | | Deposit date: | 2008-11-13 | | Release date: | 2008-12-09 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Tetrahydroisoquinoline Amide Substituted Phenyl Pyrazoles as Selective Bcl-2 Inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4P93
 
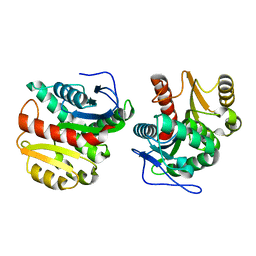 | | Structure of Dienelactone Hydrolase at 1.85 A resolution crystallised in the C2 space group | | Descriptor: | Carboxymethylenebutenolidase | | Authors: | Porter, J.L, Carr, P.D, Collyer, C.A, Ollis, D.L. | | Deposit date: | 2014-04-02 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallization of dienelactone hydrolase in two space groups: structural changes caused by crystal packing.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
4P92
 
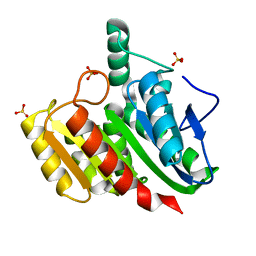 | | Crystal structure of dienelactone hydrolase C123S mutant at 1.65 A resolution | | Descriptor: | Carboxymethylenebutenolidase, SULFATE ION | | Authors: | Porter, J.L, Carr, P.D, Collyer, C.A, Ollis, D.L. | | Deposit date: | 2014-04-02 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystallization of dienelactone hydrolase in two space groups: structural changes caused by crystal packing.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
4U2B
 
 | |
4U2E
 
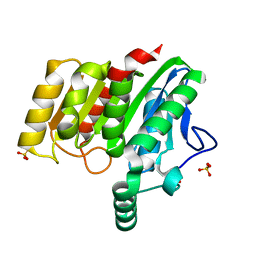 | | Crystal structure of dienelactone hydrolase S-3 variant (Q35H, F38L, Q110L, C123S, Y137C, Y145C, N154D, E199G, S208G, G211D and K234N) at 1.70 A resolution | | Descriptor: | Carboxymethylenebutenolidase, SULFATE ION | | Authors: | Porter, J.L, Collyer, C.A, Ollis, D.L. | | Deposit date: | 2014-07-16 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Directed evolution of new and improved enzyme functions using an evolutionary intermediate and multidirectional search.
Acs Chem.Biol., 10, 2015
|
|
4U2F
 
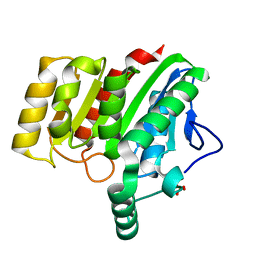 | | Crystal structure of dienelactone hydrolase B-1 variant (Q35H, F38L, Y64H, Q110L, C123S, Y137C, Y145C, N154D, E199G, S208G and G211D) at 1.80 A resolution | | Descriptor: | Carboxymethylenebutenolidase, SULFATE ION | | Authors: | Porter, J.L, Collyer, C.A, Ollis, D.L. | | Deposit date: | 2014-07-16 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Directed evolution of new and improved enzyme functions using an evolutionary intermediate and multidirectional search.
Acs Chem.Biol., 10, 2015
|
|
4U2D
 
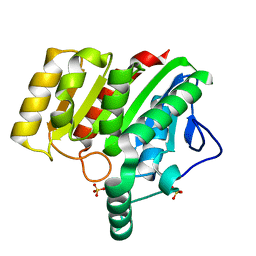 | | Crystal structure of dienelactone hydrolase S-2 variant (Q35H, F38L, Q110L, C123S, Y137C, Y145C, N154D, E199G, S208G and G211D) at 1.67 A resolution | | Descriptor: | Carboxymethylenebutenolidase, SULFATE ION | | Authors: | Porter, J.L, Collyer, C.A, Ollis, D.L. | | Deposit date: | 2014-07-16 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Directed evolution of new and improved enzyme functions using an evolutionary intermediate and multidirectional search.
Acs Chem.Biol., 10, 2015
|
|
4U2G
 
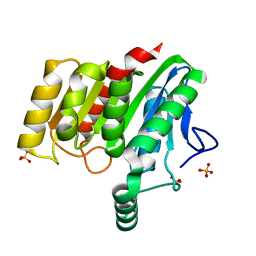 | | Crystal structure of dienelactone hydrolase B-4 variant (Q35H, F38L, Y64H, Q76L, Q110L, C123S, Y137C, A141V, Y145C, N154D, E199G, S208G, G211D, S233G and 237Q) at 1.80 A resolution | | Descriptor: | Carboxymethylenebutenolidase, SULFATE ION | | Authors: | Porter, J.L, Collyer, C.A, Ollis, D.L. | | Deposit date: | 2014-07-16 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Directed evolution of new and improved enzyme functions using an evolutionary intermediate and multidirectional search.
Acs Chem.Biol., 10, 2015
|
|
4U2C
 
 | | Crystal structure of dienelactone hydrolase A-6 variant (S7T, A24V, Q35H, F38L, Q110L, C123S, Y145C, E199G and S208G) at 1.95 A resolution | | Descriptor: | Carboxymethylenebutenolidase, SULFATE ION | | Authors: | Porter, J.L, Collyer, C.A, Ollis, D.L. | | Deposit date: | 2014-07-16 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Directed evolution of new and improved enzyme functions using an evolutionary intermediate and multidirectional search.
Acs Chem.Biol., 10, 2015
|
|
1AT0
 
 | | 17-kDA fragment of hedgehog C-terminal autoprocessing domain | | Descriptor: | 17-HEDGEHOG | | Authors: | Hall, T.M.T, Porter, J.A, Young, K.E, Koonin, E.V, Beachy, P.A, Leahy, D.J. | | Deposit date: | 1997-08-15 | | Release date: | 1997-11-12 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a Hedgehog autoprocessing domain: homology between Hedgehog and self-splicing proteins.
Cell(Cambridge,Mass.), 91, 1997
|
|
6C99
 
 | | Crystal structure of FcRn bound to UCB-303 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, CITRIC ACID, ... | | Authors: | Fox III, D, Abendroth, J, Porter, J, Deboves, H. | | Deposit date: | 2018-01-25 | | Release date: | 2018-05-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insight into small molecule binding to the neonatal Fc receptor by X-ray crystallography and 100 kHz magic-angle-spinning NMR.
PLoS Biol., 16, 2018
|
|
1VHH
 
 | | A POTENTIAL CATALYTIC SITE WITHIN THE AMINO-TERMINAL SIGNALLING DOMAIN OF SONIC HEDGEHOG | | Descriptor: | SONIC HEDGEHOG, SULFATE ION, ZINC ION | | Authors: | Hall, T.M.T, Porter, J.A, Beachy, P.A, Leahy, D.J. | | Deposit date: | 1995-10-03 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A potential catalytic site revealed by the 1.7-A crystal structure of the amino-terminal signalling domain of Sonic hedgehog.
Nature, 378, 1995
|
|
3RFN
 
 | | Epitope backbone grafting by computational design for improved presentation of linear epitopes on scaffold proteins | | Descriptor: | BB_1wnu_001, ZINC ION | | Authors: | Azoitei, M.L, Ban, Y.A, Julien, J.P, Bryson, S, Schroeter, A, Kalyuzhniy, O, Porter, J.R, Adachi, Y, Baker, D, Szabo, E, Pai, E.F, Schief, W.R. | | Deposit date: | 2011-04-06 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Computational design of high-affinity epitope scaffolds by backbone grafting of a linear epitope.
J.Mol.Biol., 415, 2012
|
|
5FO5
 
 | | Structure of the DNA-binding domain of Escherichia coli methionine biosynthesis regulator MetR | | Descriptor: | 1,2-ETHANEDIOL, HTH-TYPE TRANSCRIPTIONAL REGULATOR METR, MAGNESIUM ION | | Authors: | Punekar, A.S, Porter, J, Urbanowski, M.L, Stauffer, G.V, Carr, S.B, Phillips, S.E. | | Deposit date: | 2015-11-18 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural Basis for DNA Recognition by the Transcription Regulator Metr.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
3RHU
 
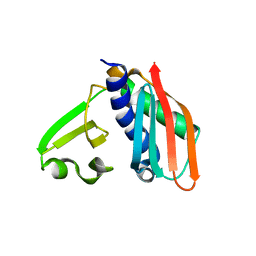 | | Epitope backbone grafting by computational design for improved presentation of linear epitopes on scaffold proteins | | Descriptor: | SC_1wnu | | Authors: | Azoitei, M.L, Ban, Y.A, Julien, J.P, Bryson, S, Schroeter, A, Kalyuzhniy, O, Porter, J.R, Adachi, Y, Baker, D, Szabo, E, Pai, E.F, Schief, W.R. | | Deposit date: | 2011-04-12 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Computational design of high-affinity epitope scaffolds by backbone grafting of a linear epitope.
J.Mol.Biol., 415, 2012
|
|
3RI0
 
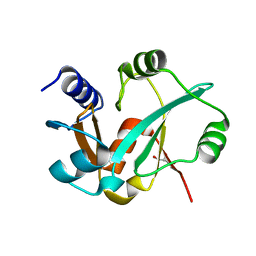 | | Epitope backbone grafting by computational design for improved presentation of linear epitopes on scaffold proteins | | Descriptor: | BB_2cx5_001, GLYCEROL, SULFATE ION | | Authors: | Azoitei, M.L, Ban, Y.A, Julien, J.P, Bryson, S, Schroeter, A, Kalyuzhniy, O, Porter, J.R, Adachi, Y, Baker, D, Szabo, E, Pai, E.F, Schief, W.R. | | Deposit date: | 2011-04-12 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Computational design of high-affinity epitope scaffolds by backbone grafting of a linear epitope.
J.Mol.Biol., 415, 2012
|
|
3RIJ
 
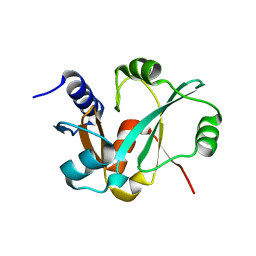 | | Epitope backbone grafting by computational design for improved presentation of linear epitopes on scaffold proteins | | Descriptor: | GLYCEROL, SC_2cx5 | | Authors: | Azoitei, M.L, Ban, Y.A, Julien, J.P, Bryson, S, Schroeter, A, Kalyuzhniy, O, Porter, J.R, Adachi, Y, Baker, D, Szabo, E, Pai, E.F, Schief, W.R. | | Deposit date: | 2011-04-13 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Computational design of high-affinity epitope scaffolds by backbone grafting of a linear epitope.
J.Mol.Biol., 415, 2012
|
|
