3L28
 
 | | Crystal structure of Zaire Ebola VP35 interferon inhibitory domain K339A mutant | | Descriptor: | CHLORIDE ION, Polymerase cofactor VP35, SODIUM ION, ... | | Authors: | Leung, D.W, Prins, K.C, Borek, D.M, Farahbakhsh, M, Tufariello, J.M, Ramanan, P, Nix, J.C, Helgeson, L.A, Otwinowski, Z, Honzatko, R.B, Basler, C.F, Amarasinghe, G.K. | | Deposit date: | 2009-12-14 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3L27
 
 | | Crystal structure of Zaire Ebola VP35 interferon inhibitory domain R312A mutant | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Leung, D.W, Prins, K.C, Borek, D.M, Farahbakhsh, M, Tufariello, J.M, Ramanan, P, Nix, J.C, Helgeson, L.A, Otwinowski, Z, Honzatko, R.B, Basler, C.F, Amarasinghe, G.K. | | Deposit date: | 2009-12-14 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3L26
 
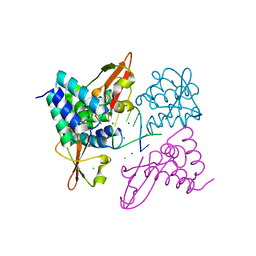 | | Crystal structure of Zaire Ebola VP35 interferon inhibitory domain bound to 8 bp dsRNA | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Polymerase cofactor VP35, ... | | Authors: | Leung, D.W, Prins, K.C, Borek, D.M, Farahbakhsh, M, Tufariello, J.M, Ramanan, P, Nix, J.C, Helgeson, L.A, Otwinowski, Z, Honzatko, R.B, Basler, C.F, Amarasinghe, G.K. | | Deposit date: | 2009-12-14 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35.
Nat.Struct.Mol.Biol., 17, 2010
|
|
7M7W
 
 | | Antibodies to the SARS-CoV-2 receptor-binding domain that maximize breadth and resistance to viral escape | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Monoclonal antibody S2H97 Fab heavy chain, Monoclonal antibody S2H97 Fab light chain, ... | | Authors: | Snell, G, Czudnochowski, N, Croll, T.I, Nix, J.C, Corti, D, Cameroni, E, Pinto, D, Beltramello, M. | | Deposit date: | 2021-03-29 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape.
Nature, 597, 2021
|
|
5TPY
 
 | | Crystal structure of an exonuclease resistant RNA from Zika virus | | Descriptor: | HEXANE-1,6-DIOL, MAGNESIUM ION, RNA (71-MER) | | Authors: | Akiyama, B.M, Laurence, H.M, Massey, A.R, Costantino, D.A, Xie, X, Yang, Y, Shi, P.-Y, Nix, J.C, Beckham, J.D, Kieft, J.S. | | Deposit date: | 2016-10-21 | | Release date: | 2016-12-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Zika virus produces noncoding RNAs using a multi-pseudoknot structure that confounds a cellular exonuclease.
Science, 354, 2016
|
|
7UOF
 
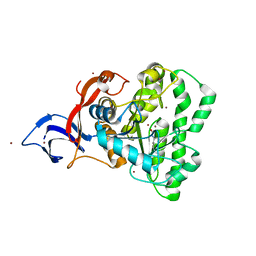 | | Dihydroorotase from M. jannaschii | | Descriptor: | Dihydroorotase, ZINC ION | | Authors: | Vitali, J, Nix, J.C, Newman, H.E, Colaneri, M.J. | | Deposit date: | 2022-04-12 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Methanococcus jannaschii dihydroorotase.
Proteins, 91, 2023
|
|
6OP7
 
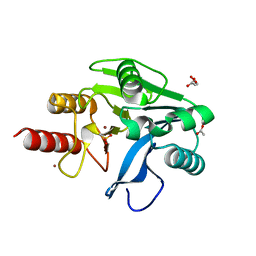 | | Structure of oxidized VIM-20 | | Descriptor: | ACETATE ION, Metallo-beta-lactamase VIM-20, ZINC ION | | Authors: | Page, R.C, Shurina, B.A, Montgomery, J.S, Orischak, M.G, Nix, J.C. | | Deposit date: | 2019-04-24 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | A Single Salt Bridge in VIM-20 Increases Protein Stability and Antibiotic Resistance under Low-Zinc Conditions.
Mbio, 10, 2019
|
|
3S9K
 
 | | Crystal structure of the Itk SH2 domain. | | Descriptor: | CITRIC ACID, Tyrosine-protein kinase ITK/TSK | | Authors: | Joseph, R.E, Ginder, N.D, Hoy, J.A, Nix, J.C, Fulton, B.D, Honzatko, R.B, Andreotti, A.H. | | Deposit date: | 2011-06-01 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.354 Å) | | Cite: | Structure of the interleukin-2 tyrosine kinase Src homology 2 domain; comparison between X-ray and NMR-derived structures.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
2OXQ
 
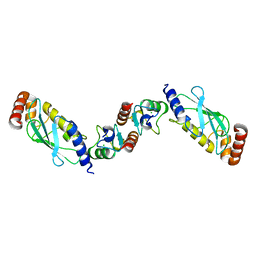 | | Structure of the UbcH5 :CHIP U-box complex | | Descriptor: | CHLORIDE ION, STIP1 homology and U-Box containing protein 1, Ubiquitin-conjugating enzyme E2D 1 | | Authors: | Xu, Z, Nix, J.C, Devlin, K.I, Misra, S. | | Deposit date: | 2007-02-20 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Interactions between the quality control ubiquitin ligase CHIP and ubiquitin conjugating enzymes.
Bmc Struct.Biol., 8, 2008
|
|
7N0E
 
 | |
7JX3
 
 | | Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab domain of monoclonal antibody S2H14, Heavy chain of Fab domain of monoclonal antibody S304, ... | | Authors: | Snell, G, Czudnochowski, N, Rosen, L.E, Nix, J.C, Corti, D, Veesler, D, Park, Y.J, Walls, A.C, Tortorici, M.A, Cameroni, E, Pinto, D, Beltramello, M, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-08-26 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7M98
 
 | | ATAD2 bromodomain complexed with histone H4K5ac (res 1-10) ligand | | Descriptor: | ATPase family AAA domain-containing protein 2, Histone H4 | | Authors: | Malone, K.L, Phillips, M, Nix, J.C, Glass, K.C. | | Deposit date: | 2021-03-30 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Coordination of Di-Acetylated Histone Ligands by the ATAD2 Bromodomain.
Int J Mol Sci, 22, 2021
|
|
3QIL
 
 | |
3FGH
 
 | | Human mitochondrial transcription factor A box B | | Descriptor: | CADMIUM ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Gangelhoff, T.A, Mungalachetty, P, Nix, J, Churchill, M.E.A. | | Deposit date: | 2008-12-06 | | Release date: | 2009-04-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural analysis and DNA binding of the HMG domains of the human mitochondrial transcription factor A
Nucleic Acids Res., 37, 2009
|
|
5F16
 
 | | CTA-modified hen egg-white lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | McGlone, C, Nix, J.C, Page, R.C. | | Deposit date: | 2015-11-30 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Investigating the Impact of Polymer Functional Groups on the Stability and Activity of Lysozyme-Polymer Conjugates.
Biomacromolecules, 17, 2016
|
|
5F14
 
 | | Structure of native hen egg-white lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | McGlone, C, Nix, J.C, Page, R.C. | | Deposit date: | 2015-11-30 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.148 Å) | | Cite: | Investigating the Impact of Polymer Functional Groups on the Stability and Activity of Lysozyme-Polymer Conjugates.
Biomacromolecules, 17, 2016
|
|
3CQX
 
 | | Chaperone Complex | | Descriptor: | BAG family molecular chaperone regulator 2, Heat shock cognate 71 kDa protein, SODIUM ION, ... | | Authors: | Xu, Z, Nix, J.C, Misra, S. | | Deposit date: | 2008-04-03 | | Release date: | 2008-11-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of nucleotide exchange and client binding by the Hsp70 cochaperone Bag2
Nat.Struct.Mol.Biol., 15, 2008
|
|
6PMT
 
 | |
3QQR
 
 | | Crystal structure of Parasponia hemoglobin; Differential Heme Coordination is Linked to Quaternary Structure | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Non-legume hemoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kakar, S, Sturms, R, Savage, A, Nix, J.C, Dispirito, A, Hargrove, M.S. | | Deposit date: | 2011-02-16 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structures of Parasponia and Trema hemoglobins: differential heme coordination is linked to quaternary structure.
Biochemistry, 50, 2011
|
|
7RNJ
 
 | | S2P6 Fab fragment bound to the SARS-CoV/SARS-CoV-2 spike stem helix peptide | | Descriptor: | Monoclonal antibody S2P6 Fab heavy chain, Monoclonal antibody S2P6 Fab light chain, SULFATE ION, ... | | Authors: | Snell, G, Czudnochowski, N, Croll, T.I, Nix, J.C, Corti, D, Cameroni, E, Pinto, D, Beltramello, M, Sauer, M.M, Veesler, D. | | Deposit date: | 2021-07-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Broad betacoronavirus neutralization by a stem helix-specific human antibody.
Science, 373, 2021
|
|
6OP6
 
 | | Structure of VIM-20 in the reduced state | | Descriptor: | Metallo-beta-lactamase VIM-20, SODIUM ION, ZINC ION | | Authors: | Page, R.C, Shurina, B.A, Montgomery, J.S, Orischak, M.G, Nix, J.C. | | Deposit date: | 2019-04-24 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A Single Salt Bridge in VIM-20 Increases Protein Stability and Antibiotic Resistance under Low-Zinc Conditions.
Mbio, 10, 2019
|
|
3D0T
 
 | |
6NHK
 
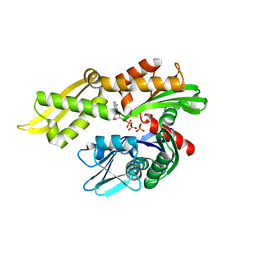 | | Mortalin nucleotide binding domain in the ADP-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Page, R.C, Moseng, M.A, Nix, J.C. | | Deposit date: | 2018-12-23 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.777 Å) | | Cite: | Biophysical Consequences of EVEN-PLUS Syndrome Mutations for the Function of Mortalin.
J.Phys.Chem.B, 123, 2019
|
|
6MJ0
 
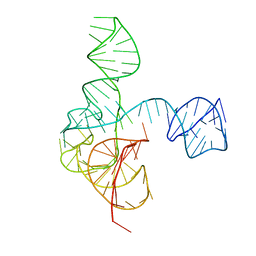 | | Crystal structure of the complete turnip yellow mosaic virus 3'UTR | | Descriptor: | RNA (101-MER) | | Authors: | Hartwick, E.W, Costantino, D.A, MacFadden, A, Nix, J.C, Tian, S, Das, R, Kieft, J.S. | | Deposit date: | 2018-09-20 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Ribosome-induced RNA conformational changes in a viral 3'-UTR sense and regulate translation levels.
Nat Commun, 9, 2018
|
|
3FKE
 
 | | Structure of the Ebola VP35 Interferon Inhibitory Domain | | Descriptor: | Polymerase cofactor VP35 | | Authors: | Amarasinghe, G.K, Leung, D.W, Ginder, N.D, Honzatko, R.B, Nix, J, Basler, C.F, Fulton, D.B. | | Deposit date: | 2008-12-16 | | Release date: | 2009-01-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the Ebola VP35 interferon inhibitory domain.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
