7N1S
 
 | | Crystal Structure Analysis of Xac Nucleotide Pyrophosphatase/Phosphodiesterase | | Descriptor: | Phosphodiesterase-nucleotide pyrophosphatase, TETRAETHYLENE GLYCOL, ZINC ION | | Authors: | Fernandez, D, Li, L, Brown, J.A, Carozza, J.A. | | Deposit date: | 2021-05-28 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ENPP1's regulation of extracellular cGAMP is a ubiquitous mechanism of attenuating STING signaling.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6VRJ
 
 | |
5XMI
 
 | | Cryo-EM Structure of the ATP-bound VPS4 mutant-E233Q hexamer (masked) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Vacuolar protein sorting-associated protein 4 | | Authors: | Sun, S, Li, L, Yang, F, Wang, X, Fan, F, Li, X, Wang, H, Sui, S. | | Deposit date: | 2017-05-15 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of the ATP-bound Vps4(E233Q) hexamer and its complex with Vta1 at near-atomic resolution
Nat Commun, 8, 2017
|
|
4IVT
 
 | | Crystal structure of BACE1 with its inhibitor | | Descriptor: | Beta-secretase 1, N-{N-[4-(acetylamino)-3,5-dichlorobenzyl]carbamimidoyl}-2-(1H-indol-1-yl)acetamide, SULFATE ION | | Authors: | Chen, T.T, Li, L, Chen, W.Y, Xu, Y.C. | | Deposit date: | 2013-01-23 | | Release date: | 2013-11-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Virtual screening and structure-based discovery of indole acylguanidines as potent beta-secretase (BACE1) inhibitors
Molecules, 18, 2013
|
|
4IVS
 
 | | Crystal structure of BACE1 with its inhibitor | | Descriptor: | Beta-secretase 1, N-{N-[4-(acetylamino)-3,5-dichlorobenzyl]carbamimidoyl}-2-(6-cyano-1H-indol-1-yl)acetamide | | Authors: | Chen, T.T, Li, L, Chen, W.Y, Xu, Y.C. | | Deposit date: | 2013-01-23 | | Release date: | 2013-11-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.636 Å) | | Cite: | Virtual screening and structure-based discovery of indole acylguanidines as potent beta-secretase (BACE1) inhibitors
Molecules, 18, 2013
|
|
4LUQ
 
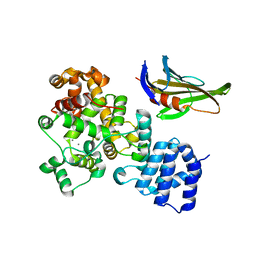 | |
3MAZ
 
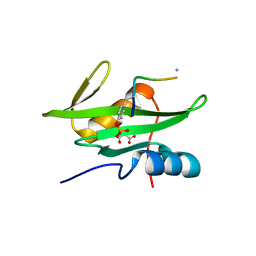 | | Crystal Structure of the Human BRDG1/STAP-1 SH2 Domain in Complex with the NTAL pTyr136 Peptide | | Descriptor: | CheD family protein, MALONATE ION, Signal-transducing adaptor protein 1 | | Authors: | Kaneko, T, Huang, H, Zhao, B, Li, L, Liu, H, Voss, C.K, Wu, C, Schiller, M.R, Li, S.S. | | Deposit date: | 2010-03-24 | | Release date: | 2010-05-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Loops govern SH2 domain specificity by controlling access to binding pockets.
Sci.Signal., 3, 2010
|
|
3II9
 
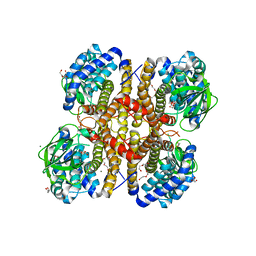 | | Crystal structure of glutaryl-coa dehydrogenase from Burkholderia pseudomallei at 1.73 Angstrom | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Glutaryl-CoA dehydrogenase, ... | | Authors: | Ismagilov, R.F, Li, L, Du, W.B, Staker, B, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-07-31 | | Release date: | 2009-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | User-loaded SlipChip for equipment-free multiplexed nanoliter-scale experiments.
J.Am.Chem.Soc., 132, 2010
|
|
3G7F
 
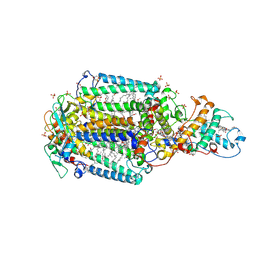 | | Crystal structure of Blastochloris viridis heterodimer mutant reaction center | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Ponomarenko, N.S, Li, L, Tereshko, V, Ismagilov, R.F, Norris Jr, J.R. | | Deposit date: | 2009-02-09 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and spectropotentiometric analysis of Blastochloris viridis heterodimer mutant reaction center
Biochim.Biophys.Acta, 1788, 2009
|
|
6CFF
 
 | | Stimulator of Interferon Genes Human | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, Stimulator of interferon genes protein | | Authors: | Fernandez, D, Li, L, Ergun, S.L. | | Deposit date: | 2018-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | STING Polymer Structure Reveals Mechanisms for Activation, Hyperactivation, and Inhibition.
Cell, 178, 2019
|
|
2LH8
 
 | | Syrian hamster prion protein with thiamine | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, Major prion protein | | Authors: | Perez-Pineiro, R, Bjorndahl, T.C, Berjanskii, M, Hau, D, Li, L, Huang, A, Lee, R, Gibbs, E, Ladner, C, Wei Dong, Y, Abera, A, Cashman, N.R, Wishart, D. | | Deposit date: | 2011-08-05 | | Release date: | 2011-09-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The prion protein binds thiamine.
Febs J., 278, 2011
|
|
2L6I
 
 | |
4F8Y
 
 | |
4FQB
 
 | |
4FCO
 
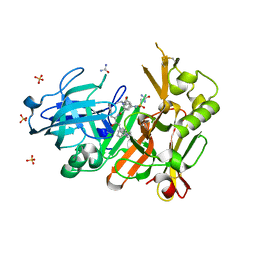 | | Crystal structure of bace1 with its inhibitor | | Descriptor: | Beta-secretase 1, N-[(2S,3R)-4-{[2-(1-benzylpiperidin-4-yl)ethyl]amino}-3-hydroxy-1-phenylbutan-2-yl]-5-[methyl(methylsulfonyl)amino]-N'-[(1R)-1-phenylethyl]benzene-1,3-dicarboxamide, SULFATE ION, ... | | Authors: | Chen, T.T, Chen, W.Y, Li, L, Xu, Y.C. | | Deposit date: | 2012-05-25 | | Release date: | 2013-05-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Flexibility of the Flap in the Active Site of BACE1 as Revealed by Crystal Structures and MD simulations
To be Published, 2012
|
|
3RT3
 
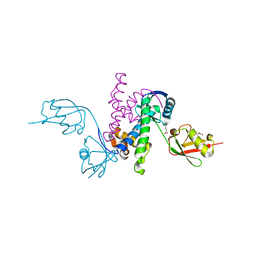 | |
3F9G
 
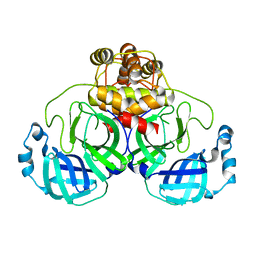 | |
3F9F
 
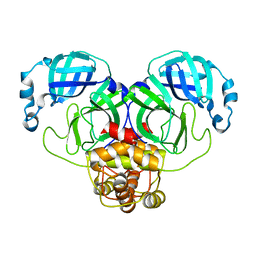 | |
3F9H
 
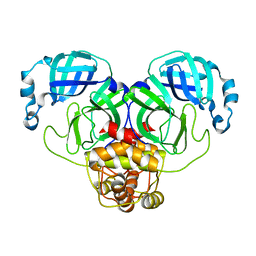 | |
3C6R
 
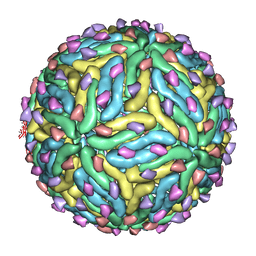 | | Low pH Immature Dengue Virus | | Descriptor: | Envelope protein, Peptide pr | | Authors: | Yu, I, Zhang, W, Holdway, H.A, Li, L, Kostyuchenko, V.A, Chipman, P.R, Kuhn, R.J, Rossmann, M.G, Chen, J. | | Deposit date: | 2008-02-05 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (25 Å) | | Cite: | Structure of the immature dengue virus at low pH primes proteolytic maturation
Science, 319, 2008
|
|
3F9E
 
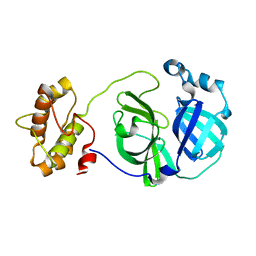 | |
7XHA
 
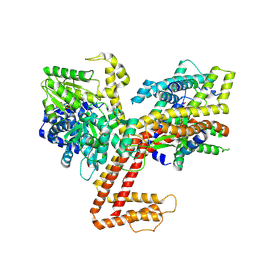 | |
7XHB
 
 | |
6M3B
 
 | | hAPC-c25k23 Fab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Vitamin K-dependent protein C heavy chain, Vitamin K-dependent protein C light chain, ... | | Authors: | Wang, X, Li, L, Zhao, X, Egner, U. | | Deposit date: | 2020-03-03 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Targeted inhibition of activated protein C by a non-active-site inhibitory antibody to treat hemophilia.
Nat Commun, 11, 2020
|
|
6LO8
 
 | | Cryo-EM structure of the TIM22 complex from yeast | | Descriptor: | Mitochondrial import inner membrane translocase subunit TIM10, Mitochondrial import inner membrane translocase subunit TIM12, Mitochondrial import inner membrane translocase subunit TIM18, ... | | Authors: | Zhang, Y, Zhou, X, Wu, X, Li, L. | | Deposit date: | 2020-01-04 | | Release date: | 2020-09-30 | | Last modified: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | Structure of the mitochondrial TIM22 complex from yeast.
Cell Res., 31, 2021
|
|
