7AA9
 
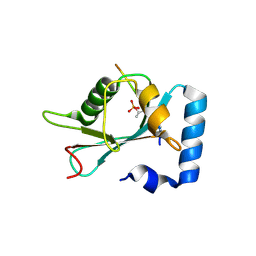 | | Structure of SCOC pT13/pT15 LIR motif bound to GABARAPL1 | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein-like 1, pT13/PT15 SCOC LIR | | Authors: | Lee, R, Mouilleron, S, Wirth, M, Zhang, W, O Reilly, N, Dhira, J, Tooze, S. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Phosphorylation of the LIR Domain of SCOC Modulates ATG8 Binding Affinity and Specificity.
J.Mol.Biol., 433, 2021
|
|
7AA8
 
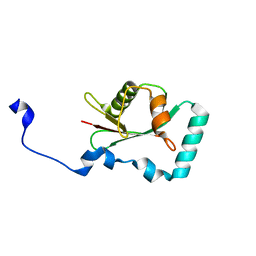 | | Structure of SCOC LIR bound to GABARAP | | Descriptor: | Chimera made of SCOC (6-23) + linker (GS) + GABARAP,Gamma-aminobutyric acid receptor-associated protein | | Authors: | Lee, R, Mouilleron, S, Wirth, M, Zhang, W, O Reilly, N, Dhira, J, Tooze, S. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Phosphorylation of the LIR Domain of SCOC Modulates ATG8 Binding Affinity and Specificity.
J.Mol.Biol., 433, 2021
|
|
7AA7
 
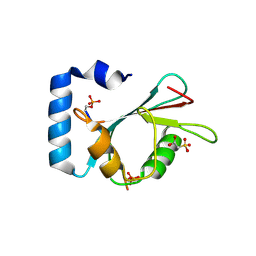 | | Structure of SCOC pS12/pS18 LIR motif bound to GABARAPL1 | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein-like 1, pS12/pS18 SCOC LIR, sulfoacetic acid | | Authors: | Lee, R, Mouilleron, S, Wirth, M, Zhang, W, O Reilly, N, Dhira, J, Tooze, S. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Phosphorylation of the LIR Domain of SCOC Modulates ATG8 Binding Affinity and Specificity.
J.Mol.Biol., 433, 2021
|
|
8E7P
 
 | | Staphylococcus aureus ClpP in complex with compound 3421 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (5R,6S,9aS)-N-benzyl-6-[(4-hydroxyphenyl)methyl]-8-[(naphthalen-1-yl)methyl]-4,7-dioxohexahydro-2H-pyrazino[1,2-a]pyrimidine-1(6H)-carboxamide, ATP-dependent Clp protease proteolytic subunit | | Authors: | Lee, R.E, Griffith, E.C. | | Deposit date: | 2022-08-24 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Development of a high throughput and site specific, fluorescent polarization assay to screen for activators of Caseinolytic Protease P leads to the discovery of synthetically tractable new activator class
To Be Published
|
|
8E71
 
 | | Staphylococcus aureus ClpP in complex with compound 3471 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (5S,6S,9aS)-N-[(4-fluorophenyl)methyl]-6-methyl-8-[(naphthalen-1-yl)methyl]-4,7-dioxohexahydro-2H-pyrazino[1,2-a]pyrimidine-1(6H)-carboxamide, ATP-dependent Clp protease proteolytic subunit | | Authors: | Lee, R.E, Griffith, E.C. | | Deposit date: | 2022-08-23 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Development of a high throughput and site specific, fluorescent polarization assay to screen for activators of Caseinolytic Protease P leads to the discovery of synthetically tractable new activator class
To Be Published
|
|
8EF8
 
 | | Staphylococcus aureus ClpP Y63W in complex with compound 3471 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (5S,6S,9aS)-N-[(4-fluorophenyl)methyl]-6-methyl-8-[(naphthalen-1-yl)methyl]-4,7-dioxohexahydro-2H-pyrazino[1,2-a]pyrimidine-1(6H)-carboxamide, ATP-dependent Clp protease proteolytic subunit | | Authors: | Lee, R.E, Griffith, E.C. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Development of a high throughput and site specific, fluorescent polarization assay to screen for activators of Caseinolytic Protease P leads to the discovery of synthetically tractable new activator class
To Be Published
|
|
5W18
 
 | | Staphylococcus aureus ClpP in complex with (S)-N-((2R,6S,8aS,14aS,20S,23aS)-2,6-dimethyl-5,8,14,19,23-pentaoxooctadecahydro-1H,5H,14H,19H-pyrido[2,1-i]dipyrrolo[2,1-c:2',1'-l][1]oxa[4,7,10,13]tetraazacyclohexadecin-20-yl)-3-phenyl-2-(3-phenylureido)propanamide | | Descriptor: | 9V7-PHE-SER-PRO-YCP-ALA-MP8, ATP-dependent Clp protease proteolytic subunit | | Authors: | Lee, R.E, Griffith, E.C. | | Deposit date: | 2017-06-02 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Ureadepsipeptides as ClpP Activators.
Acs Infect Dis., 2019
|
|
6CFD
 
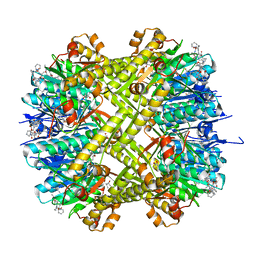 | | ADEP4 bound to E. faecium ClpP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ATP-dependent Clp protease proteolytic subunit, N-[(6aS,12S,15aS,17R,21R,23aS)-17,21-dimethyl-6,11,15,20,23-pentaoxooctadecahydro-2H,6H,11H,15H-pyrido[2,1-i]dipyrrolo[2,1-c:2',1'-l][1,4,7,10,13]oxatetraazacyclohexadecin-12-yl]-3,5-difluoro-Nalpha-[(2E)-hept-2-enoyl]-L-phenylalaninamide | | Authors: | Lee, R.E, Griffith, E.C. | | Deposit date: | 2018-02-14 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | In VivoandIn VitroEffects of a ClpP-Activating Antibiotic against Vancomycin-Resistant Enterococci.
Antimicrob. Agents Chemother., 62, 2018
|
|
6N80
 
 | | S. aureus ClpP bound to anti-4a | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, N-[(1R)-1-borono-3-methylbutyl]-N~2~-(2-chloro-4-methoxybenzene-1-carbonyl)-L-leucinamide | | Authors: | Lee, R.E, Griffith, E.C. | | Deposit date: | 2018-11-28 | | Release date: | 2019-06-26 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | De Novo Design of Boron-Based Peptidomimetics as Potent Inhibitors of Human ClpP in the Presence of Human ClpX.
J.Med.Chem., 62, 2019
|
|
7JLS
 
 | | RV3666c bound to tripeptide | | Descriptor: | Peptide SER-VAL-ALA, Probable periplasmic dipeptide-binding lipoprotein DppA | | Authors: | Lee, R.E, Griffith, E.C, Miller, D.J. | | Deposit date: | 2020-07-30 | | Release date: | 2021-06-09 | | Last modified: | 2021-12-29 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Biophysical analysis of the Mycobacteria tuberculosis peptide binding protein DppA reveals a stringent peptide binding pocket.
Tuberculosis (Edinb), 132, 2021
|
|
8TXA
 
 | | Apo Structure of (N1G37) tRNA Methyltransferase from Mycobacterium marinum | | Descriptor: | tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Balsamo, A, Bruno, C, Edele, C, Fabian, T, Jannotta, R, Lee, R, Schryver, D, Warsaw, J, Warsaw, L, Stojanoff, V, Battaile, K, Perez, A, Bolen, R. | | Deposit date: | 2023-08-23 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Apo Structure of (N1G37) tRNA Methyltransferase from Mycobacterium marinum
To Be Published
|
|
6ZEF
 
 | | Structure of PP1(7-300) bound to Phactr1 (516-580) at pH 5.25 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
6ZEG
 
 | | Structure of PP1-IRSp53 chimera [PP1(7-304) + linker (G/S)x9 + IRSp53(449-465)] bound to Phactr1 (516-580) | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE, MANGANESE (II) ION, ... | | Authors: | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
6ZEE
 
 | | Structure of PP1(7-300) bound to Phactr1 (507-580) at pH8.4 | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE, GLYCEROL, ... | | Authors: | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
6ZEI
 
 | | Structure of PP1-IRSp53 S455E chimera [PP1(7-304) + linker (G/S)x9 + IRSp53(449-465)] bound to Phactr1 (516-580) | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
6ZEH
 
 | | Structure of PP1-spectrin alpha II chimera [PP1(7-304) + linker (G/S)x9 + spectrin alpha II (1025-1039)] bound to Phactr1 (516-580) | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, Phosphatase and actin regulator, ... | | Authors: | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
6PKA
 
 | |
6PMD
 
 | |
5N2M
 
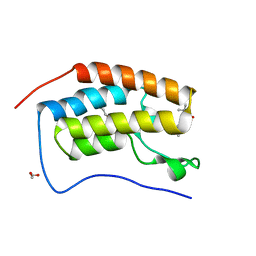 | | Crystal structure of the first bromodomain of human BRD4 in complex with a tetrahydroquinoline analogue | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, propan-2-yl ~{N}-[(2~{S},4~{R})-6-(3-acetamidophenyl)-1-ethanoyl-2-methyl-3,4-dihydro-2~{H}-quinolin-4-yl]carbamate | | Authors: | Tallant, C, Slavish, P.J, Siejka, P, Bharatham, N, Shadrick, W.R, Chai, S, Young, B.M, Boyd, V.A, Heroven, C, Wiggers, H.J, Picaud, S, Fedorov, O, Krojer, T, Chen, T, Lee, R.E, Guy, R.K, Shelat, A.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-02-07 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure of the first bromodomain of human BRD4 in complex with a tetrahydroquinoline analogue
To Be Published
|
|
5N2L
 
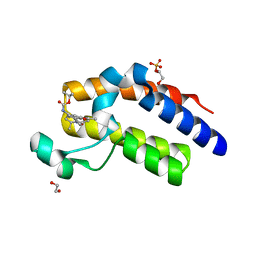 | | Crystal structure of the second bromodomain of human BRD2 in complex with a tetrahydroquinoline analogue | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 2, SULFATE ION, ... | | Authors: | Tallant, C, Slavish, P.J, Siejka, P, Bharatham, N, Shadrick, W.R, Chai, S, Young, B.M, Boyd, V.A, Heroven, C, Wiggers, H.J, Picaud, S, Fedorov, O, Krojer, T, Chen, T, Lee, R.E, Guy, R.K, Shelat, A.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-02-07 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of the second bromodomain of human BRD2 in complex with a tetrahydroquinoline analogue
To Be Published
|
|
5M9Q
 
 | | Human angiogenin PD variant R95Q | | Descriptor: | Angiogenin, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Bradshaw, W.J, Rehman, S, Pham, T.T.K, Thiyagarajan, N, Lee, R.L, Subramanian, V, Acharya, K.R. | | Deposit date: | 2016-11-02 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural insights into human angiogenin variants implicated in Parkinson's disease and Amyotrophic Lateral Sclerosis.
Sci Rep, 7, 2017
|
|
5M9T
 
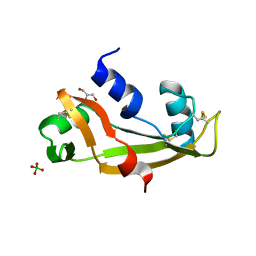 | | Human angiogenin ALS variant H114R | | Descriptor: | Angiogenin, BORATE ION, GLYCEROL | | Authors: | Bradshaw, W.J, Rehman, S, Pham, T.T.K, Thiyagarajan, N, Lee, R.L, Subramanian, V, Acharya, K.R. | | Deposit date: | 2016-11-02 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into human angiogenin variants implicated in Parkinson's disease and Amyotrophic Lateral Sclerosis.
Sci Rep, 7, 2017
|
|
5M9G
 
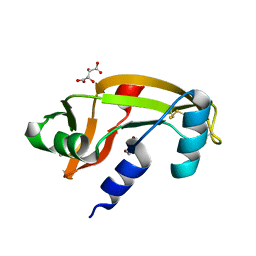 | | Human angiogenin PD variant K54R | | Descriptor: | Angiogenin, D(-)-TARTARIC ACID, DI(HYDROXYETHYL)ETHER | | Authors: | Bradshaw, W.J, Rehman, S, Pham, T.T.K, Thiyagarajan, N, Lee, R.L, Subramanian, V, Acharya, K.R. | | Deposit date: | 2016-11-01 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural insights into human angiogenin variants implicated in Parkinson's disease and Amyotrophic Lateral Sclerosis.
Sci Rep, 7, 2017
|
|
5M9S
 
 | | Human angiogenin ALS variant V103I | | Descriptor: | Angiogenin, D(-)-TARTARIC ACID | | Authors: | Bradshaw, W.J, Rehman, S, Pham, T.T.K, Thiyagarajan, N, Lee, R.L, Subramanian, V, Acharya, K.R. | | Deposit date: | 2016-11-02 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insights into human angiogenin variants implicated in Parkinson's disease and Amyotrophic Lateral Sclerosis.
Sci Rep, 7, 2017
|
|
5M9P
 
 | | Human angiogenin ALS variant T80S | | Descriptor: | Angiogenin, SULFATE ION | | Authors: | Bradshaw, W.J, Rehman, S, Pham, T.T.K, Thiyagarajan, N, Lee, R.L, Subramanian, V, Acharya, K.R. | | Deposit date: | 2016-11-02 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into human angiogenin variants implicated in Parkinson's disease and Amyotrophic Lateral Sclerosis.
Sci Rep, 7, 2017
|
|
