1UOT
 
 | | HUMAN CD55 DOMAINS 3 & 4 | | Descriptor: | COMPLEMENT DECAY-ACCELERATING FACTOR | | Authors: | Williams, P, Chaudhry, Y, Goodfellow, I.G, Billington, J, Spiller, B, Evans, D.J, Lea, S.M. | | Deposit date: | 2003-09-23 | | Release date: | 2003-09-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mapping Cd55 Function. The Structure of Two Pathogen-Binding Domains at 1.7 A
J.Biol.Chem., 278, 2003
|
|
8GL6
 
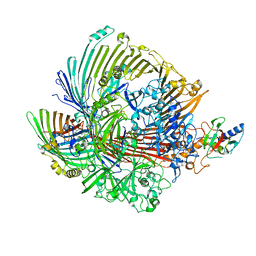 | |
8GLK
 
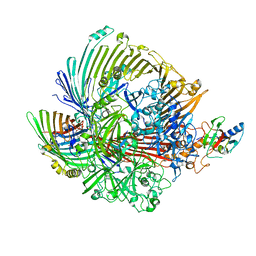 | |
8GLN
 
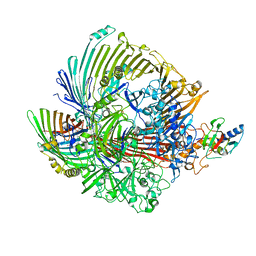 | |
8GLM
 
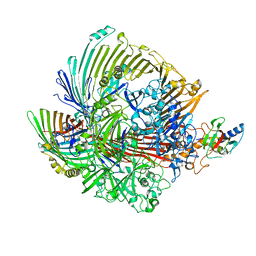 | |
8GL8
 
 | |
8GLJ
 
 | |
5NQY
 
 | | Structure of a fHbp(V1.4):PorA(P1.16) chimera. Fusion at fHbp position 309. | | Descriptor: | Factor H binding protein variant B16_001,Major outer membrane protein P.IA,Factor H binding protein variant B16_001 | | Authors: | Johnson, S, Hollingshead, S, Lea, S.M, Tang, C.M. | | Deposit date: | 2017-04-21 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based design of chimeric antigens for multivalent protein vaccines.
Nat Commun, 9, 2018
|
|
5NQZ
 
 | | Structure of a fHbp(V1.1):PorA(P1.16) chimera. Fusion at fHbp position 309. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Factor H binding protein,Major outer membrane protein P.IA,Factor H binding protein, ... | | Authors: | Johnson, S, Hollingshead, S, Lea, S.M, Tang, C.M. | | Deposit date: | 2017-04-21 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structure-based design of chimeric antigens for multivalent protein vaccines.
Nat Commun, 9, 2018
|
|
5NQX
 
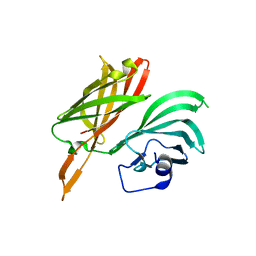 | | Structure of a fHbp(V1.1):PorA(P1.16) chimera. Fusion at fHbp position 294. | | Descriptor: | Factor H binding protein,Major outer membrane protein P.IA,Factor H binding protein | | Authors: | Johnson, S, Jongerius, I, Lea, S.M, Tang, C.M. | | Deposit date: | 2017-04-21 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.66 Å) | | Cite: | Structure-based design of chimeric antigens for multivalent protein vaccines.
Nat Commun, 9, 2018
|
|
5NQP
 
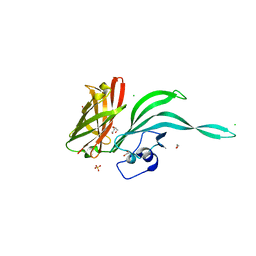 | | Structure of a fHbp(V1.4):PorA(P1.16) chimera. Fusion at fHbp position 151. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Factor H binding protein variant B16_001,Major outer membrane protein P.IA,Factor H binding protein variant B16_001, ... | | Authors: | Johnson, S, Hollingshead, S, Lea, S.M, Tang, C.M. | | Deposit date: | 2017-04-20 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structure-based design of chimeric antigens for multivalent protein vaccines.
Nat Commun, 9, 2018
|
|
8HCN
 
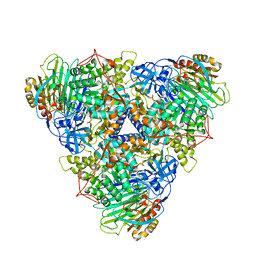 | | CryoEM Structure of Klebsiella pneumoniae UreD/urease complex | | Descriptor: | Urease accessory protein UreD, Urease subunit alpha, Urease subunit beta, ... | | Authors: | Nim, Y.S, Fong, I.Y.H, Deme, J, Tsang, K.L, Caesar, J, Johnson, S, Wong, K.B, Lea, S.M. | | Deposit date: | 2022-11-02 | | Release date: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Delivering a toxic metal to the active site of urease.
Sci Adv, 9, 2023
|
|
8HC1
 
 | | CryoEM structure of Helicobacter pylori UreFD/urease complex | | Descriptor: | Urease accessory protein UreF, Urease accessory protein UreH, Urease subunit alpha, ... | | Authors: | Nim, Y.S, Fong, I.Y.H, Deme, J, Tsang, K.L, Caesar, J, Johnson, S, Wong, K.B, Lea, S.M. | | Deposit date: | 2022-11-01 | | Release date: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Delivering a toxic metal to the active site of urease.
Sci Adv, 9, 2023
|
|
2J0O
 
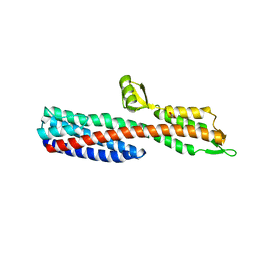 | | Shigella Flexneri IpaD | | Descriptor: | GLYCEROL, INVASIN IPAD | | Authors: | Johnson, S, Roversi, P, Lea, S.M. | | Deposit date: | 2006-08-04 | | Release date: | 2006-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Self-chaperoning of the type III secretion system needle tip proteins IpaD and BipD.
J. Biol. Chem., 282, 2007
|
|
6H3J
 
 | |
6H3I
 
 | | Structural snapshots of the Type 9 protein translocon | | Descriptor: | Peptidyl-prolyl cis-trans isomerase, PorV, Protein involved in gliding motility SprA | | Authors: | Deme, J.C, Lea, S.M. | | Deposit date: | 2018-07-18 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Type 9 secretion system structures reveal a new protein transport mechanism.
Nature, 564, 2018
|
|
2IXR
 
 | | BipD of Burkholderia Pseudomallei | | Descriptor: | MEMBRANE ANTIGEN | | Authors: | Roversi, P, Johnson, S, Field, T, Deane, J.E, Galyov, E.E, Lea, S.M. | | Deposit date: | 2006-07-10 | | Release date: | 2006-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Self-Chaperoning of the Type III Secretion System Needle Tip Proteins Ipad and Bipd.
J.Biol.Chem., 282, 2007
|
|
2J0N
 
 | |
2J9T
 
 | | BipD of Burkholderia Pseudomallei | | Descriptor: | BORIC ACID, CITRATE ANION, MEMBRANE ANTIGEN | | Authors: | Johnson, S, Roversi, P, Field, T, Deane, J.E, Galyov, E, Lea, S.M. | | Deposit date: | 2006-11-16 | | Release date: | 2006-11-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Self-Chaperoning of the Type III Secretion System Needle Tip Proteins Ipad and Bipd.
J.Biol.Chem., 282, 2007
|
|
1H8T
 
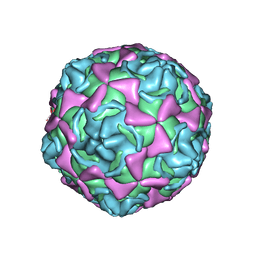 | | Echovirus 11 | | Descriptor: | 12-AMINO-DODECANOIC ACID, ECHOVIRUS 11 COAT PROTEIN VP1, ECHOVIRUS 11 COAT PROTEIN VP2, ... | | Authors: | Stuart, A, McKee, T, Williams, P.A, Harley, C, Stuart, D.I, Brown, T.D.K, Lea, S.M. | | Deposit date: | 2001-02-15 | | Release date: | 2002-07-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Determination of the Structure of a Decay Accelerating Factor-Binding Clinical Isolate of Echovirus 11 Allows Mapping of Mutants with Altered Receptor Requirements for Infection
J.Virol., 76, 2002
|
|
8SA3
 
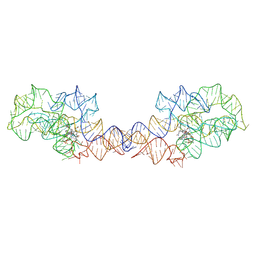 | | Adenosylcobalamin-bound riboswitch dimer, form 2 | | Descriptor: | Adenosylcobalamin, adenosylcobalamin riboswitch form 2 | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8SA6
 
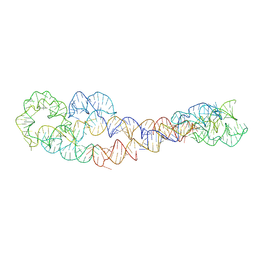 | | apo form of adenosylcobalamin riboswitch dimer | | Descriptor: | apo form of adenosylcobalamin riboswitch dimer | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8SA5
 
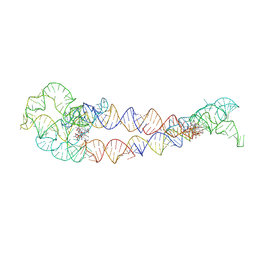 | | Adenosylcobalamin-bound riboswitch dimer, form 4 | | Descriptor: | Adenosylcobalamin, adenosylcobalamin riboswitch form 4 | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8SA4
 
 | | Adenosylcobalamin-bound riboswitch dimer, form 3 | | Descriptor: | Adenosylcobalamin, adenosylcobalamin riboswitch form 3 | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8SA2
 
 | | Adenosylcobalamin-bound riboswitch dimer, form 1 | | Descriptor: | Adenosylcobalamin, adenosylcobalamin riboswitch form 1 | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
