5WPZ
 
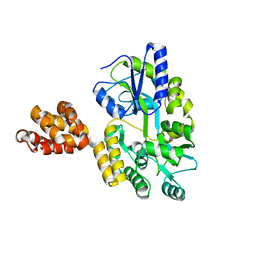 | | Crystal structure of MNDA PYD with MBP tag | | Descriptor: | MBP-hMNDA-PYD, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Jin, T.C, Xiao, T.S. | | Deposit date: | 2016-11-22 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of an expression system to enhance MBP-mediated crystallization
Sci Rep, 7, 2017
|
|
2RE9
 
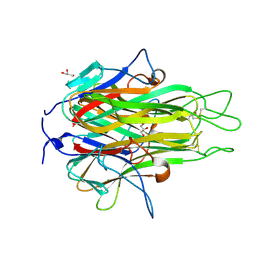 | | Crystal structure of TL1A at 2.1 A | | Descriptor: | GLYCEROL, MAGNESIUM ION, TNF superfamily ligand TL1A | | Authors: | Jin, T.C, Guo, F, Kim, S, Howard, A.J, Zhang, Y.Z. | | Deposit date: | 2007-09-25 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray crystal structure of TNF ligand family member TL1A at 2.1 A.
Biochem.Biophys.Res.Commun., 364, 2007
|
|
3GIX
 
 | |
3VD8
 
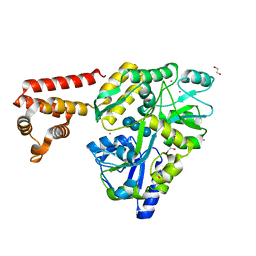 | | Crystal structure of human AIM2 PYD domain with MBP fusion | | Descriptor: | 1,2-ETHANEDIOL, Maltose-binding periplasmic protein, Interferon-inducible protein AIM2, ... | | Authors: | Jin, T.C, Perry, A, Smith, P, Xiao, T.S. | | Deposit date: | 2012-01-04 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.0685 Å) | | Cite: | Structure of the Absent in Melanoma 2 (AIM2) Pyrin Domain Provides Insights into the Mechanisms of AIM2 Autoinhibition and Inflammasome Assembly.
J.Biol.Chem., 288, 2013
|
|
5WQ6
 
 | | Crystal Structure of hMNDA-PYD with MBP tag | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, MBP tagged hMNDA-PYD, ... | | Authors: | Jin, T.C, Xiao, T.S. | | Deposit date: | 2016-11-23 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.648 Å) | | Cite: | Design of an expression system to enhance MBP-mediated crystallization
Sci Rep, 7, 2017
|
|
3FZ3
 
 | | Crystal Structure of almond Pru1 protein | | Descriptor: | CALCIUM ION, Prunin, SODIUM ION | | Authors: | Jin, T.C, Zhang, Y.Z. | | Deposit date: | 2009-01-23 | | Release date: | 2009-11-10 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of prunin-1, a major component of the almond (Prunus dulcis) allergen amandin.
J.Agric.Food Chem., 57, 2009
|
|
4IN0
 
 | |
4LEJ
 
 | | Crystal Structure of the Korean pine (Pinus koraiensis) vicilin | | Descriptor: | COPPER (II) ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Jin, T.C, Wang, Y, Zhang, Y.Z. | | Deposit date: | 2013-06-25 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Crystal Structure of Korean Pine ( Pinus koraiensis ) 7S Seed Storage Protein with Copper Ligands.
J.Agric.Food Chem., 62, 2014
|
|
3RN2
 
 | | Structural Basis of Cytosolic DNA Recognition by Innate Immune Receptors | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*TP*CP*AP*AP*AP*GP*AP*TP*CP*TP*TP*TP*GP*AP*TP*GP*G)-3'), Interferon-inducible protein AIM2 | | Authors: | Jin, T.C, Xiao, T. | | Deposit date: | 2011-04-21 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structures of the HIN Domain:DNA Complexes Reveal Ligand Binding and Activation Mechanisms of the AIM2 Inflammasome and IFI16 Receptor.
Immunity, 36, 2012
|
|
3RLN
 
 | |
3RLO
 
 | | Structural Basis of Cytosolic DNA Recognition by Innate Receptors | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Gamma-interferon-inducible protein 16 | | Authors: | Jin, T.C, Xiao, T. | | Deposit date: | 2011-04-19 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the HIN Domain:DNA Complexes Reveal Ligand Binding and Activation Mechanisms of the AIM2 Inflammasome and IFI16 Receptor.
Immunity, 36, 2012
|
|
3RN5
 
 | | Structural basis of cytosolic DNA recognition by innate immune receptors | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*TP*CP*AP*AP*AP*GP*AP*GP*AP*GP*AP*AP*AP*GP*AP*G)-3'), DNA (5'-D(*GP*CP*TP*CP*TP*TP*TP*CP*TP*CP*TP*CP*TP*TP*TP*GP*AP*TP*G)-3'), ... | | Authors: | Jin, T.C, Xiao, T. | | Deposit date: | 2011-04-21 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the HIN Domain:DNA Complexes Reveal Ligand Binding and Activation Mechanisms of the AIM2 Inflammasome and IFI16 Receptor.
Immunity, 36, 2012
|
|
3RNU
 
 | | Structural Basis of Cytosolic DNA Sensing by Innate Immune Receptors | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*CP*AP*TP*CP*AP*AP*AP*GP*AP*GP*AP*GP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*CP*TP*CP*TP*TP*TP*GP*AP*TP*GP*GP*CP*C)-3'), ... | | Authors: | Jin, T.C, Xiao, T. | | Deposit date: | 2011-04-22 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structures of the HIN Domain:DNA Complexes Reveal Ligand Binding and Activation Mechanisms of the AIM2 Inflammasome and IFI16 Receptor.
Immunity, 36, 2012
|
|
5H7Q
 
 | | Crystal structure of human MNDA PYD domain with MBP tag | | Descriptor: | ACETATE ION, MNDA PYD domain with MBP tag, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Jin, T.C, Xiao, T.S. | | Deposit date: | 2016-11-20 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Design of an expression system to enhance MBP-mediated crystallization
Sci Rep, 7, 2017
|
|
5H7N
 
 | | Crystal structure of human NLRP12-PYD with a MBP tag | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NLRP12-PYD with MBP tag, ... | | Authors: | Jin, T.C, Xiao, T.S. | | Deposit date: | 2016-11-19 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Design of an expression system to enhance MBP-mediated crystallization
Sci Rep, 7, 2017
|
|
5H6I
 
 | | Crystal Structure of GBS CAMP Factor | | Descriptor: | CHLORIDE ION, Protein B, SULFATE ION | | Authors: | Jin, T.C, Brefo-Mensah, E.K. | | Deposit date: | 2016-11-13 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of theStreptococcus agalactiaeCAMP factor provides insights into its membrane-permeabilizing activity.
J.Biol.Chem., 293, 2018
|
|
6JLC
 
 | |
2O0O
 
 | | Crystal structure of TL1A | | Descriptor: | MAGNESIUM ION, TNF superfamily ligand TL1A | | Authors: | Jin, T.C, Kim, S, Guo, F, Howard, A.J, Zhang, Y.Z. | | Deposit date: | 2006-11-27 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray crystal structure of TNF ligand family member TL1A at 2.1A.
Biochem.Biophys.Res.Commun., 364, 2007
|
|
2NML
 
 | | Crystal structure of HEF2/ERH at 1.55 A resolution | | Descriptor: | Enhancer of rudimentary homolog | | Authors: | Jin, T.C, Guo, F, Serebriiskii, I.G, Howard, A.J, Zhang, Y.Z. | | Deposit date: | 2006-10-21 | | Release date: | 2006-10-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A 1.55 A resolution X-ray crystal structure of HEF2/ERH and insights into its transcriptional and cell-cycle interaction networks.
Proteins, 68, 2007
|
|
8IWF
 
 | | Aspergillus niger Rha-2 and pNPR | | Descriptor: | (2S,3R,4R,5R,6S)-2-methyl-6-[4-[oxidanyl(oxidanylidene)-$l^4-azanyl]phenoxy]oxane-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, L.J, Li, L, Wang, M.H, Jiang, Z.D, Zhu, Y.B, Jin, T.C, Ni, H. | | Deposit date: | 2023-03-29 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Crystal structure and catalytic function of alpha-L-rhamnosidase from Aspergillus niger
To Be Published
|
|
8IWD
 
 | | Aspergillus niger Rha-2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, L.J, Li, L, Wang, M.H, Jiang, Z.D, Zhu, Y.B, Jin, T.C, Ni, H. | | Deposit date: | 2023-03-29 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal structure and catalytic function of alpha-L-rhamnosidase from Aspergillus niger
To Be Published
|
|
7X4I
 
 | |
8YE4
 
 | | The complex of TCR NYN-I and HLA-A24 bound to SARS-CoV-2 Spike448-456 peptide NYNYLYRLF | | Descriptor: | Beta-2-microglobulin, MHC class I antigen precusor, Spike protein S1, ... | | Authors: | Deng, S.S, Jin, T.C, Xu, Z.H, Wang, M.H. | | Deposit date: | 2024-02-21 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into immune escape at killer T cell epitope by SARS-CoV-2 Spike Y453F variants.
J.Biol.Chem., 300, 2024
|
|
8ZV9
 
 | | Complex structure of HLA2402 with recognizing SARS-CoV-2 Y453F epitope NYNYLFRLF | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, MHC class I antigen, ... | | Authors: | Deng, S.S, Jin, T.C, Xu, Z.H, Wang, M.H. | | Deposit date: | 2024-06-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into immune escape at killer T cell epitope by SARS-CoV-2 Spike Y453F variants.
J.Biol.Chem., 300, 2024
|
|
8WCG
 
 | |
