4JC8
 
 | |
4KMO
 
 | | Crystal Structure of the Vps33-Vps16 HOPS subcomplex from Chaetomium thermophilum | | Descriptor: | Putative vacuolar protein sorting-associated protein, SULFATE ION, Small conjugating protein ligase-like protein | | Authors: | Baker, R.W, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2013-05-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of the Sec1/Munc18 (SM) Protein Vps33, Alone and Bound to the Homotypic Fusion and Vacuolar Protein Sorting (HOPS) Subunit Vps16*
Plos One, 8, 2013
|
|
2F1W
 
 | | Crystal structure of the TRAF-like domain of HAUSP/USP7 | | Descriptor: | CALCIUM ION, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Hu, M, Gu, L, Jeffrey, P.D, Shi, Y. | | Deposit date: | 2005-11-15 | | Release date: | 2006-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis of Competitive Recognition of p53 and MDM2 by HAUSP/USP7: Implications for the Regulation of the p53-MDM2 Pathway.
Plos Biol., 4, 2006
|
|
4L9O
 
 | | Crystal Structure of the Sec13-Sec16 blade-inserted complex from Pichia pastoris | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | McMahon, C, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2013-06-18 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Sec16 influences transitional ER sites by regulating rather than organizing COPII.
Mol Biol Cell, 24, 2013
|
|
2F1S
 
 | | Crystal Structure of a Viral FLIP MC159 | | Descriptor: | Viral CASP8 and FADD-like apoptosis regulator | | Authors: | Li, F.-Y, Jeffrey, P.D, Yu, J.W, Shi, Y. | | Deposit date: | 2005-11-15 | | Release date: | 2005-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of a Viral FLIP: INSIGHTS INTO FLIP-MEDIATED INHIBITION OF DEATH RECEPTOR SIGNALING.
J.Biol.Chem., 281, 2006
|
|
2F1Y
 
 | |
2NYM
 
 | | Crystal Structure of Protein Phosphatase 2A (PP2A) with C-terminus truncated catalytic subunit | | Descriptor: | MANGANESE (II) ION, Protein phosphatase 2, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform, ... | | Authors: | Chen, Y, Xing, Y, Xu, Y, Chao, Y, Lin, Z, Jeffrey, P.D, Shi, Y. | | Deposit date: | 2006-11-21 | | Release date: | 2006-12-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of the Protein Phosphatase 2A Holoenzyme.
Cell(Cambridge,Mass.), 127, 2006
|
|
4I5K
 
 | |
2F1Z
 
 | | Crystal structure of HAUSP | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Hu, M, Gu, L, Jeffrey, P.D, Shi, Y. | | Deposit date: | 2005-11-15 | | Release date: | 2006-02-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis of Competitive Recognition of p53 and MDM2 by HAUSP/USP7: Implications for the Regulation of the p53-MDM2 Pathway.
Plos Biol., 4, 2006
|
|
2F1X
 
 | |
4M9R
 
 | | Crystal structure of CED-3 | | Descriptor: | Cell death protein 3 | | Authors: | Xu, Y, Jeffrey, P.D, Shi, Y.G. | | Deposit date: | 2013-08-15 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.656 Å) | | Cite: | Mechanistic insights into CED-4-mediated activation of CED-3
Genes Dev., 27, 2013
|
|
2HV6
 
 | |
2IE3
 
 | | Structure of the Protein Phosphatase 2A Core Enzyme Bound to Tumor-inducing Toxins | | Descriptor: | MANGANESE (II) ION, Protein Phosphatase 2, regulatory subunit A (PR 65), ... | | Authors: | Xing, Y, Xu, Y, Chen, Y, Jeffrey, P.D, Chao, Y, Shi, Y. | | Deposit date: | 2006-09-17 | | Release date: | 2006-11-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Protein Phosphatase 2A Core Enzyme Bound to Tumor-Inducing Toxins
Cell(Cambridge,Mass.), 127, 2006
|
|
2IE4
 
 | | Structure of the Protein Phosphatase 2A Core Enzyme Bound to okadaic acid | | Descriptor: | MANGANESE (II) ION, OKADAIC ACID, Protein Phosphatase 2, ... | | Authors: | Xing, Y, Xu, Y, Chen, Y, Jeffrey, P.D, Chao, Y, Shi, Y. | | Deposit date: | 2006-09-17 | | Release date: | 2006-11-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Protein Phosphatase 2A Core Enzyme Bound to Tumor-Inducing Toxins
Cell(Cambridge,Mass.), 127, 2006
|
|
3DW8
 
 | | Structure of a Protein Phosphatase 2A Holoenzyme with B55 subunit | | Descriptor: | MANGANESE (II) ION, Serine/threonine-protein phosphatase 2A 55 kDa regulatory subunit B alpha isoform, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, ... | | Authors: | Xu, Y, Chen, Y, Zhang, P, Jeffrey, P.D, Shi, Y. | | Deposit date: | 2008-07-21 | | Release date: | 2008-10-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of a protein phosphatase 2A holoenzyme: insights into B55-mediated Tau dephosphorylation.
Mol.Cell, 31, 2008
|
|
1MYK
 
 | | CRYSTAL STRUCTURE, FOLDING, AND OPERATOR BINDING OF THE HYPERSTABLE ARC REPRESSOR MUTANT PL8 | | Descriptor: | ARC REPRESSOR | | Authors: | Schildbach, J.F, Milla, M.E, Jeffrey, P.D, Raumann, B.E, Sauer, R.T. | | Deposit date: | 1994-10-12 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure, folding, and operator binding of the hyperstable Arc repressor mutant PL8.
Biochemistry, 34, 1995
|
|
1SHX
 
 | | Ephrin A5 ligand structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ephrin-A5 | | Authors: | Himanen, J.P, Barton, W.A, Nikolov, D.B, Jeffrey, P.D. | | Deposit date: | 2004-02-26 | | Release date: | 2005-04-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three distinct molecular surfaces in ephrin-A5 are essential for a functional interaction with EphA3.
J.Biol.Chem., 280, 2005
|
|
3CEV
 
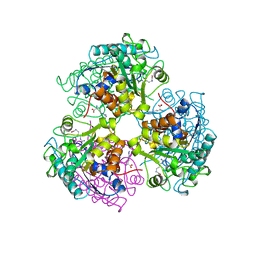 | | ARGINASE FROM BACILLUS CALDEVELOX, COMPLEXED WITH L-ARGININE | | Descriptor: | ARGININE, MANGANESE (II) ION, PROTEIN (ARGINASE) | | Authors: | Bewley, M.C, Jeffrey, P.D, Patchett, M.L, Kanyo, Z.F, Baker, E.N. | | Deposit date: | 1999-03-15 | | Release date: | 1999-04-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of Bacillus caldovelox arginase in complex with substrate and inhibitors reveal new insights into activation, inhibition and catalysis in the arginase superfamily.
Structure Fold.Des., 7, 1999
|
|
3HQT
 
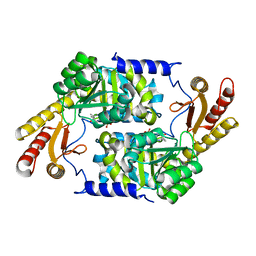 | | PLP-Dependent Acyl-CoA Transferase CqsA | | Descriptor: | CAI-1 autoinducer synthase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Kelly, R.C, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2009-06-08 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Vibrio cholerae quorum-sensing autoinducer CAI-1: analysis of the biosynthetic enzyme CqsA.
Nat.Chem.Biol., 5, 2009
|
|
3ETU
 
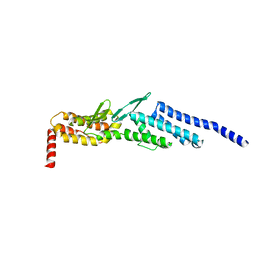 | | Crystal structure of yeast Dsl1p | | Descriptor: | Protein transport protein DSL1 | | Authors: | Ren, Y, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2008-10-08 | | Release date: | 2009-01-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural characterization of Tip20p and Dsl1p, subunits of the Dsl1p vesicle tethering complex.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3FHN
 
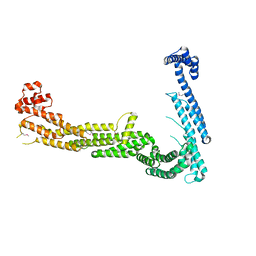 | | Structure of Tip20p | | Descriptor: | Protein transport protein TIP20 | | Authors: | Tripathi, A, Ren, Y, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2008-12-09 | | Release date: | 2009-01-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural characterization of Tip20p and Dsl1p, subunits of the Dsl1p vesicle tethering complex.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3HR0
 
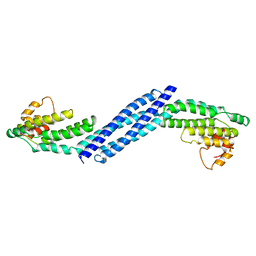 | | Crystal structure of Homo sapiens Conserved Oligomeric Golgi subunit 4 | | Descriptor: | CoG4 | | Authors: | Richardson, B.C, Ungar, D, Nakamura, A, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2009-06-08 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for a human glycosylation disorder caused by mutation of the COG4 gene.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3ETV
 
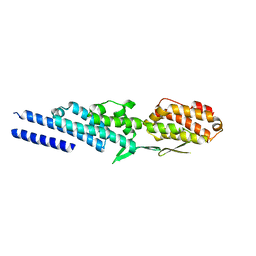 | | Crystal structure of a Tip20p-Dsl1p fusion protein | | Descriptor: | Protein transport protein TIP20, Protein transport protein DSL1 chimera | | Authors: | Ren, Y, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2008-10-08 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural characterization of Tip20p and Dsl1p, subunits of the Dsl1p vesicle tethering complex.
Nat.Struct.Mol.Biol., 16, 2009
|
|
6MYW
 
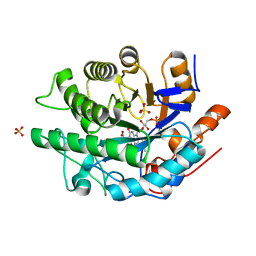 | | Gluconobacter Ene-Reductase (GluER) mutant - T36A | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Garfinkle, S.E, Jeffrey, P, Hyster, T.K. | | Deposit date: | 2018-11-02 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.157 Å) | | Cite: | Photoexcitation of flavoenzymes enables a stereoselective radical cyclization.
Science, 364, 2019
|
|
6O08
 
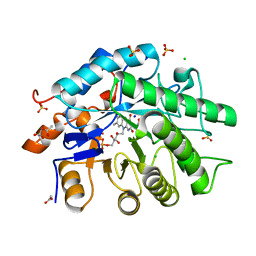 | | Gluconobacter Ene-Reductase (GluER) | | Descriptor: | ACETATE ION, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Garfinkle, S.E, Jeffrey, P, Hyster, T.K. | | Deposit date: | 2019-02-15 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Photoexcitation of flavoenzymes enables a stereoselective radical cyclization.
Science, 364, 2019
|
|
