1BIJ
 
 | | CROSSLINKED, DEOXY HUMAN HEMOGLOBIN A | | Descriptor: | BUT-2-ENEDIAL, HEMOGLOBIN A, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fernandez, E.J, Abad-Zapatero, C, Olsen, K.W. | | Deposit date: | 1998-06-18 | | Release date: | 1999-03-30 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Lysbeta(1)82-Lysbeta(2)82 crosslinked hemoglobin: a possible allosteric intermediate
J.Mol.Biol., 296, 2000
|
|
1CM9
 
 | |
3UVV
 
 | | Crystal Structure of the ligand binding domains of the thyroid receptor:retinoid X receptor complexed with 3,3',5 triiodo-L-thyronine and 9-cis retinoic acid | | Descriptor: | (9cis)-retinoic acid, 3,5,3'TRIIODOTHYRONINE, Retinoic acid receptor RXR-alpha, ... | | Authors: | Fernandez, E.J, Putcha, B.-D.K, Wright, E, Brunzelle, J.S. | | Deposit date: | 2011-11-30 | | Release date: | 2012-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis for negative cooperativity within agonist-bound TR:RXR heterodimers.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1XNX
 
 | | Crystal structure of constitutive androstane receptor | | Descriptor: | 16,17-ANDROSTENE-3-OL, constitutive androstane receptor | | Authors: | Fernandez, E. | | Deposit date: | 2004-10-05 | | Release date: | 2005-01-04 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the murine constitutive androstane receptor complexed to androstenol; a molecular basis for inverse agonism
Mol.Cell, 16, 2004
|
|
1KR3
 
 | | Crystal Structure of the Metallo beta-Lactamase from Bacteroides fragilis (CfiA) in Complex with the Tricyclic Inhibitor SB-236050. | | Descriptor: | 7,8-DIHYDROXY-1-METHOXY-3-METHYL-10-OXO-4,10-DIHYDRO-1H,3H-PYRANO[4,3-B]CHROMENE-9-CARBOXYLIC ACID, SODIUM ION, ZINC ION, ... | | Authors: | Payne, D.J, Hueso-Rodrguez, J.A, Boyd, H, Concha, N.O, Janson, C.A, Gilpin, M, Bateson, J.H, Cheever, C, Niconovich, N.L, Pearson, S, Rittenhouse, S, Tew, D, Dez, E, Prez, P, de la Fuente, J, Rees, M, Rivera-Sagredo, A. | | Deposit date: | 2002-01-08 | | Release date: | 2003-01-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of a series of tricyclic natural products as potent broad-spectrum inhibitors of metallo-beta-lactamases
ANTIMICROB.AGENTS CHEMOTHER., 46, 2002
|
|
6WBO
 
 | |
1LU1
 
 | | THE STRUCTURE OF THE DOLICHOS BIFLORUS SEED LECTIN IN COMPLEX WITH THE FORSSMAN DISACCHARIDE | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, ADENINE, CALCIUM ION, ... | | Authors: | Hamelryck, T.W, Loris, R, Bouckaert, J, Strecker, G, Imberty, A, Fernandez, E, Wyns, L, Etzler, M.E. | | Deposit date: | 1998-07-24 | | Release date: | 1998-12-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Carbohydrate binding, quaternary structure and a novel hydrophobic binding site in two legume lectin oligomers from Dolichos biflorus.
J.Mol.Biol., 286, 1999
|
|
4UOH
 
 | | Crystallographic structure of nucleoside diphosphate kinase from Litopenaeus vannamei complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Lopez-Zavala, A.A, Guevara-Hernandez, E, Stojanoff, V, Rudino-Pinera, E, Sotelo-Mundo, R.R. | | Deposit date: | 2014-06-03 | | Release date: | 2014-09-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Structure of Nucleoside Diphosphate Kinase from Pacific Shrimp (Litopenaeus Vannamei) in Binary Complexes with Purine and Pyrimidine Nucleoside Diphosphates
Acta Crystallogr.,Sect.F, 79, 2014
|
|
4OOW
 
 | | HCV NS5B polymerase with a fragment of quercetagetin | | Descriptor: | CATECHOL, RNA-directed RNA polymerase | | Authors: | Guichou, J.F, Ahmed-Belkacem, A, Rozenn, B, Nazim, N, Hernandez, E, Pallier, C, Pawlotsky, J.M. | | Deposit date: | 2014-02-04 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Inhibition of RNA binding to hepatitis C virus RNA-dependent RNA polymerase: a new mechanism for antiviral intervention.
Nucleic Acids Res., 42, 2014
|
|
6TGG
 
 | | scFv-1SM3 in complex with glycopeptide containing an sp2-imino sugar | | Descriptor: | 1,2-ETHANEDIOL, Mucin-1, ScFv_SM3, ... | | Authors: | Bermejo, I.A, Navo, C.D, Castro-Lopez, J, Samchez-Fernandez, E.M, Guerreiro, A, Avenoza, A, Busto, J.H, Garcia-Martin, F, Nishimura, S.I, Garcia-Fernandez, J.M, Ortiz-Mellet, C, Bernardes, G.J.L, Hurtado-Guerrero, R, Peregrina, J.M, Corzana, F. | | Deposit date: | 2019-11-15 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis, conformational analysis and in vivo assays of an anti-cancer vaccine that features an unnatural antigen based on an sp 2 -iminosugar fragment.
Chem Sci, 11, 2020
|
|
6BT9
 
 | | Chitinase ChiA74 from Bacillus thuringiensis | | Descriptor: | CALCIUM ION, Chitinase | | Authors: | Juarez-Hernandez, E, Brieba, L.G, Torres-Larios, A, Jimenez-Sandoval, P, Barboza-Corona, J. | | Deposit date: | 2017-12-05 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | The crystal structure of the chitinase ChiA74 of Bacillus thuringiensis has a multidomain assembly.
Sci Rep, 9, 2019
|
|
1AB6
 
 | | STRUCTURE OF CHEY MUTANT F14N, V86T | | Descriptor: | CHEMOTAXIS PROTEIN CHEY | | Authors: | Wilcock, D, Pisabarro, M.T, Lopez-Hernandez, E, Serranno, L, Coll, M. | | Deposit date: | 1997-02-04 | | Release date: | 1998-02-04 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure analysis of two CheY mutants: importance of the hydrogen-bond contribution to protein stability.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1AB5
 
 | | STRUCTURE OF CHEY MUTANT F14N, V21T | | Descriptor: | CHEY | | Authors: | Wilcock, D, Pisabarro, M.T, Lopez-Hernandez, E, Serrano, L, Coll, M. | | Deposit date: | 1997-02-04 | | Release date: | 1998-02-04 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure analysis of two CheY mutants: importance of the hydrogen-bond contribution to protein stability.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
8A3T
 
 | | S. cerevisiae APC/C-Cdh1 complex | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 11, Anaphase-promoting complex subunit 2, ... | | Authors: | Barford, D, Vazquez-Fernandez, E, Zhang, Z, Yang, J. | | Deposit date: | 2022-06-09 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of the S. cerevisiae APC/C-Cdh1 complex
To Be Published
|
|
1A15
 
 | | SDF-1ALPHA | | Descriptor: | STROMAL DERIVED FACTOR-1ALPHA, SULFATE ION | | Authors: | Dealwis, C.G, Fernandez, E.J, Lolis, E. | | Deposit date: | 1997-12-22 | | Release date: | 1998-08-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of chemically synthesized [N33A] stromal cell-derived factor 1alpha, a potent ligand for the HIV-1 "fusin" coreceptor.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
5AJL
 
 | | Sdsa sulfatase tetragonal | | Descriptor: | ALKYL SULFATASE, ZINC ION | | Authors: | De la Mora, E, Flores-Hernandez, E, Jakoncic, J, Stojanoff, V, Sanchez-Puig, N, Moreno, A. | | Deposit date: | 2015-02-25 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Sdsa Polymorph Isolation and Improvement of Their Crystal Quality Using Nonconventional Crystallization Techniques
J.Appl.Crystallogr., 48, 2015
|
|
5A23
 
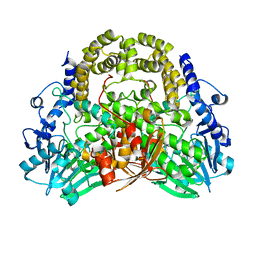 | | SdsA sulfatase triclinic form | | Descriptor: | SDS HYDROLASE SDSA1, ZINC ION | | Authors: | De la Mora, E, Flores-Hernandez, E, Jakoncik, J, Stojanoff, V, Sanchez-Puig, N, Moreno, A. | | Deposit date: | 2015-05-11 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Sdsa Polymorph Isolation and Improvement of Their Crystal Quality Using Nonconventional Crystallization Techniques
J.Appl.Crystallogr., 48, 2015
|
|
5AIJ
 
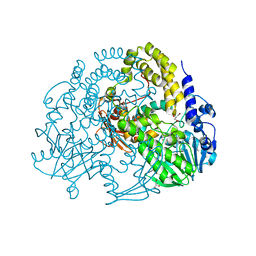 | | P. aeruginosa SdsA hexagonal polymorph | | Descriptor: | ALKYL SULFATASE, GLYCEROL, ZINC ION | | Authors: | De la Mora, E, Flores-Hernandez, E, Jakoncic, J, Stojanoff, V, Sanchez-Puig, N, Moreno, A. | | Deposit date: | 2015-02-13 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Sdsa Polymorph Isolation and Improvement of Their Crystal Quality Using Nonconventional Crystallization Techniques
J.Appl.Crystallogr., 48, 2015
|
|
1E6K
 
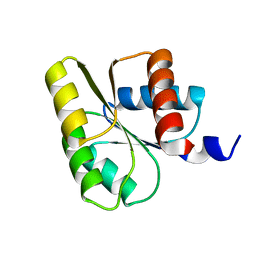 | | Two-component signal transduction system D12A mutant of CheY | | Descriptor: | Chemotaxis protein CheY | | Authors: | Sola, M, Lopez-Hernandez, E, Cronet, P, Lacroix, E, Serrano, L, Coll, M, Parraga, A. | | Deposit date: | 2000-08-18 | | Release date: | 2001-03-05 | | Last modified: | 2019-09-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Towards understanding a molecular switch mechanism: thermodynamic and crystallographic studies of the signal transduction protein CheY.
J.Mol.Biol., 303, 2000
|
|
3ZBY
 
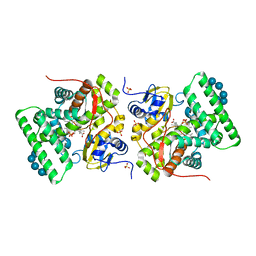 | | Ligand-free structure of CYP142 from Mycobacterium smegmatis | | Descriptor: | 1,2-ETHANEDIOL, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), P450 HEME-THIOLATE PROTEIN, ... | | Authors: | Garcia-Fernandez, E, Frank, D.J, Galan, B, Kells, P.M, Podust, L.M, Garcia, J.L, Ortiz de Montellano, P.R. | | Deposit date: | 2012-11-13 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A Highly Conserved Mycobacterial Cholesterol Catabolic Pathway.
Environ.Microbiol., 15, 2013
|
|
1E6M
 
 | | TWO-COMPONENT SIGNAL TRANSDUCTION SYSTEM D57A MUTANT OF CHEY | | Descriptor: | CHEMOTAXIS PROTEIN CHEY | | Authors: | Sola, M, Lopez-Hernandez, E, Cronet, P, Lacroix, E, Serrano, L, Coll, M, Parraga, A. | | Deposit date: | 2000-08-18 | | Release date: | 2000-09-21 | | Last modified: | 2019-09-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Towards understanding a molecular switch mechanism: thermodynamic and crystallographic studies of the signal transduction protein CheY.
J.Mol.Biol., 303, 2000
|
|
1E6L
 
 | | Two-component signal transduction system D13A mutant of CheY | | Descriptor: | Chemotaxis protein CheY | | Authors: | Sola, M, Lopez-Hernandez, E, Cronet, P, Lacroix, E, Serrano, L, Coll, M, Parraga, A. | | Deposit date: | 2000-08-18 | | Release date: | 2001-03-05 | | Last modified: | 2019-09-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Towards understanding a molecular switch mechanism: thermodynamic and crystallographic studies of the signal transduction protein CheY.
J.Mol.Biol., 303, 2000
|
|
4GNY
 
 | | Bovine beta-lactoglobulin complex with dodecyl sulfate | | Descriptor: | Beta-lactoglobulin, DODECYL SULFATE, GLYCEROL | | Authors: | Gutierrez-Magdaleno, G, Torres-Rivera, A, Garcia-Hernandez, E, Rodriguez-Romero, A. | | Deposit date: | 2012-08-17 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6367 Å) | | Cite: | Ligand-binding and self-association cooperativity of beta-lactoglobulin
J.Mol.Recognit., 26, 2013
|
|
2YOO
 
 | | Cholest-4-en-3-one bound structure of CYP142 from Mycobacterium smegmatis | | Descriptor: | (8ALPHA,9BETA)-CHOLEST-4-EN-3-ONE, MAGNESIUM ION, P450 HEME-THIOLATE PROTEIN, ... | | Authors: | Garcia-Fernandez, E, Frank, D.J, Galan, B, Kells, P.M, Podust, L.M, Garcia, J.L, Ortiz de Montellano, P.R. | | Deposit date: | 2012-10-25 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A Highly Conserved Mycobacterial Cholesterol Catabolic Pathway.
Environ.Microbiol., 15, 2013
|
|
2K1J
 
 | | Plan homeodomain finger of tumour supressor ING4 | | Descriptor: | Inhibitor of growth protein 4, ZINC ION | | Authors: | Palacios, A, Garcia, P, Padro, D, Lopez-Hernandez, E, Blanco, F.J. | | Deposit date: | 2008-03-05 | | Release date: | 2008-04-15 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure and NMR characterization of the binding to methylated histone tails of the plant homeodomain finger of the tumour suppressor ING4.
Febs Lett., 580, 2006
|
|
