2WJ2
 
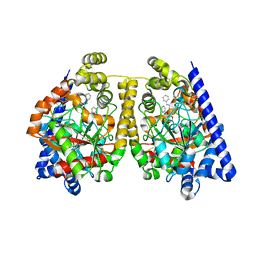 | | 3D-crystal structure of humanized-rat fatty acid amide hydrolase (FAAH) conjugated with 7-phenyl-1-(5-(pyridin-2-yl)oxazol-2-yl)heptan- 1-one, an alpha-ketooxazole | | Descriptor: | 7-phenyl-1-(5-pyridin-2-yl-1,3-oxazol-2-yl)heptane-1,1-diol, CHLORIDE ION, FATTY ACID AMIDE HYDROLASE 1 | | Authors: | Mileni, M, Garfunkle, J, DeMartino, J.K, Cravatt, B.F, Boger, D.L, Stevens, R.C. | | Deposit date: | 2009-05-19 | | Release date: | 2009-09-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Binding and Inactivation Mechanism of a Humanized Fatty Acid Amide Hydrolase by Alpha-Ketoheterocycle Inhibitors Revealed from Cocrystal Structures.
J.Am.Chem.Soc., 131, 2009
|
|
2WJ1
 
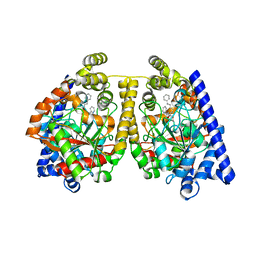 | | 3D-crystal structure of humanized-rat fatty acid amide hydrolase (FAAH) conjugated with 7-phenyl-1-(4-(pyridin-2-yl)oxazol-2-yl)heptan- 1-one, an alpha-ketooxazole | | Descriptor: | 7-phenyl-1-(4-pyridin-2-yl-1,3-oxazol-2-yl)heptane-1,1-diol, CHLORIDE ION, FATTY-ACID AMIDE HYDROLASE 1 | | Authors: | Mileni, M, Garfunkle, J, DeMartino, J.K, Cravatt, B.F, Boger, D.L, Stevens, R.C. | | Deposit date: | 2009-05-19 | | Release date: | 2009-09-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Binding and Inactivation Mechanism of a Humanized Fatty Acid Amide Hydrolase by Alpha-Ketoheterocycle Inhibitors Revealed from Cocrystal Structures.
J.Am.Chem.Soc., 131, 2009
|
|
2WHJ
 
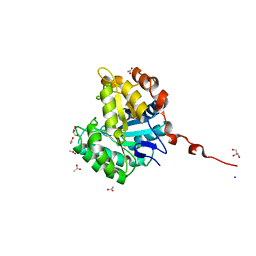 | | Understanding how diverse mannanases recognise heterogeneous substrates | | Descriptor: | ACETATE ION, BETA-MANNANASE, GLYCEROL, ... | | Authors: | Tailford, L.E, Ducros, V.M.A, Flint, J.E, Roberts, S.M, Morland, C, Zechel, D.L, Smith, N, Bjornvad, M.E, Borchert, T.V, Wilson, K.S, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2009-05-05 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Understanding How Diverse -Mannanases Recognise Heterogeneous Substrates.
Biochemistry, 48, 2009
|
|
2WYI
 
 | | Structure of the Streptococcus pyogenes family GH38 alpha-mannosidase complexed with swainsonine | | Descriptor: | 1S-8AB-OCTAHYDRO-INDOLIZIDINE-1A,2A,8B-TRIOL, 2-(2-METHOXYETHOXY)ETHANOL, ALPHA-MANNOSIDASE, ... | | Authors: | Suits, M.D.L, Zhu, Y, Taylor, E.J, Zechel, D.L, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2009-11-16 | | Release date: | 2010-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Kinetic Investigation of Streptococcus Pyogenes Family Gh38 Alpha-Mannosidase
Plos One, 5, 2010
|
|
6WER
 
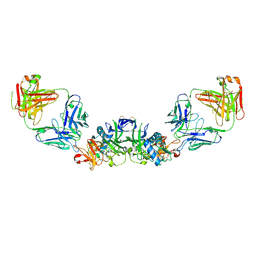 | | DENV2 NS1 in complex with neutralizing 2B7 Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2B7 Fab heavy chain, 2B7 Fab light chain, ... | | Authors: | Akey, D.L, Smith, J.L. | | Deposit date: | 2020-04-02 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.96 Å) | | Cite: | Structural basis for antibody inhibition of flavivirus NS1-triggered endothelial dysfunction.
Science, 371, 2021
|
|
2WYH
 
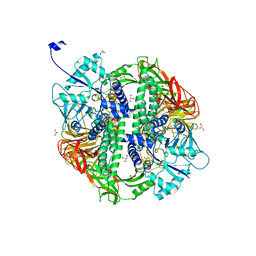 | | Structure of the Streptococcus pyogenes family GH38 alpha-mannosidase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ALPHA-MANNOSIDASE, GLYCEROL, ... | | Authors: | Suits, M.D.L, Zhu, Y, Taylor, E.J, Zechel, D.L, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2009-11-16 | | Release date: | 2010-02-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Kinetic Investigation of Streptococcus Pyogenes Family Gh38 Alpha-Mannosidase
Plos One, 5, 2010
|
|
6WEQ
 
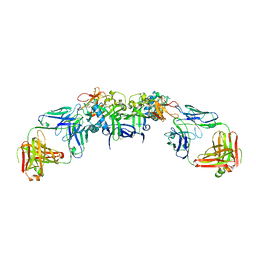 | | DENV1 NS1 in complex with neutralizing 2B7 Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2B7 Fab fragment heavy chain, 2B7 Fab fragment light chain, ... | | Authors: | Akey, D.L, Smith, J.L. | | Deposit date: | 2020-04-02 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for antibody inhibition of flavivirus NS1-triggered endothelial dysfunction.
Science, 371, 2021
|
|
2PII
 
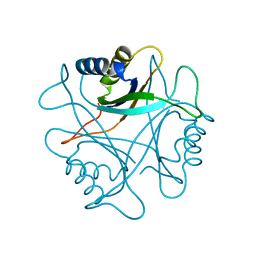 | | PII, GLNB PRODUCT | | Descriptor: | PII | | Authors: | Carr, P.D, Cheah, E, Suffolk, P.M, Ollis, D.L. | | Deposit date: | 1995-05-02 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structure of the signal transduction protein from Escherichia coli at 1.9 A.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
2ZD2
 
 | |
2ZOA
 
 | | Malonate-bound structure of the glycerophosphodiesterase from Enterobacter aerogenes (GpdQ) COLLECTED AT 1.280 ANGSTROM | | Descriptor: | FE (II) ION, MALONATE ION, Phosphohydrolase | | Authors: | Ollis, D.L, Jackson, C.J, Carr, P.D. | | Deposit date: | 2008-05-07 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Malonate-bound structure of the glycerophosphodiesterase from Enterobacter aerogenes (GpdQ) and characterization of the native Fe2+ metal-ion preference.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
2ZO9
 
 | | Malonate-bound structure of the glycerophosphodiesterase from Enterobacter aerogenes (GpdQ) and characterization of the native Fe2+ metal ion preference | | Descriptor: | FE (II) ION, MALONATE ION, Phosphohydrolase | | Authors: | Jackson, C.J, Carr, P.D, Ollis, D.L. | | Deposit date: | 2008-05-07 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Malonate-bound structure of the glycerophosphodiesterase from Enterobacter aerogenes (GpdQ) and characterization of the native Fe2+ metal-ion preference.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
3A3W
 
 | | Structure of OpdA mutant (G60A/A80V/S92A/R118Q/K185R/Q206P/D208G/I260T/G273S) with diethyl 4-methoxyphenyl phosphate bound in the active site | | Descriptor: | COBALT (II) ION, DIETHYL 4-METHOXYPHENYL PHOSPHATE, Phosphotriesterase | | Authors: | Ollis, D.L, Tawfik, D.S, Schenk, G, Jackson, C.J, Foo, J.L, Tokuriki, N, Afriat, L, Carr, P.D, Kim, H.K. | | Deposit date: | 2009-06-23 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformational sampling, catalysis, and evolution of the bacterial phosphotriesterase
Proc.Natl.Acad.Sci.USA, 2009
|
|
3A3X
 
 | | Structure of OpdA mutant (G60A/A80V/R118Q/K185R/Q206P/D208G/I260T/G273S) | | Descriptor: | COBALT (II) ION, Phosphotriesterase | | Authors: | Ollis, D.L, Tawfik, D.S, Schenk, G, Jackson, C.J, Foo, J.L, Tokuriki, N, Afriat, L, Carr, P.D, Kim, H.K. | | Deposit date: | 2009-06-23 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational sampling, catalysis, and evolution of the bacterial phosphotriesterase
Proc.Natl.Acad.Sci.USA, 2009
|
|
3A60
 
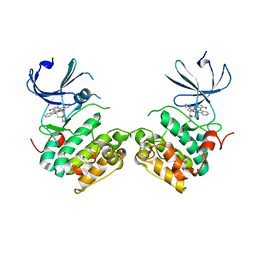 | | Crystal structure of unphosphorylated p70S6K1 (Form I) | | Descriptor: | Ribosomal protein S6 kinase beta-1, STAUROSPORINE | | Authors: | Sunami, T, Byrne, N, Diehl, R.E, Funabashi, K, Hall, D.L, Ikuta, M, Patel, S.B, Shipman, J.M, Smith, R.F, Takahashi, I, Zugay-Murphy, J, Iwasawa, Y, Lumb, K.J, Munshi, S.K, Sharma, S. | | Deposit date: | 2009-08-17 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of human p70 ribosomal S6 kinase-1 regulation by activation loop phosphorylation.
J.Biol.Chem., 285, 2010
|
|
3A4J
 
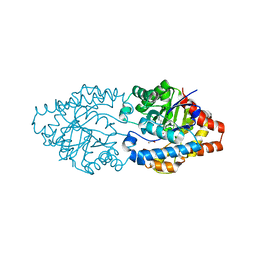 | | arPTE (K185R/D208G/N265D/T274N) | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, Phosphotriesterase | | Authors: | Foo, J.L, Jackson, C.J, Carr, P.D, Ollis, D.L. | | Deposit date: | 2009-07-07 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Conformational sampling, catalysis, and evolution of the bacterial phosphotriesterase
Proc.Natl.Acad.Sci.USA, 2009
|
|
3A61
 
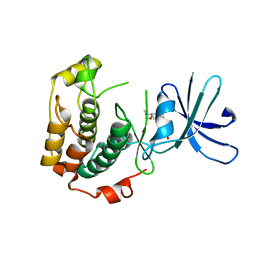 | | Crystal structure of unphosphorylated p70S6K1 (Form II) | | Descriptor: | Ribosomal protein S6 kinase beta-1, STAUROSPORINE | | Authors: | Sunami, T, Byrne, N, Diehl, R.E, Funabashi, K, Hall, D.L, Ikuta, M, Patel, S.B, Shipman, J.M, Smith, R.F, Takahashi, I, Zugay-Murphy, J, Iwasawa, Y, Lumb, K.J, Munshi, S.K, Sharma, S. | | Deposit date: | 2009-08-18 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | Structural basis of human p70 ribosomal S6 kinase-1 regulation by activation loop phosphorylation.
J.Biol.Chem., 285, 2010
|
|
3A62
 
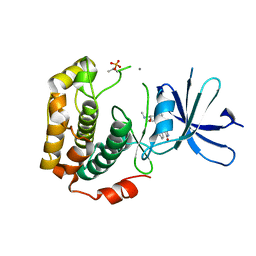 | | Crystal structure of phosphorylated p70S6K1 | | Descriptor: | MANGANESE (II) ION, Ribosomal protein S6 kinase beta-1, STAUROSPORINE | | Authors: | Sunami, T, Byrne, N, Diehl, R.E, Funabashi, K, Hall, D.L, Ikuta, M, Patel, S.B, Shipman, J.M, Smith, R.F, Takahashi, I, Zugay-Murphy, J, Iwasawa, Y, Lumb, K.J, Munshi, S.K, Sharma, S. | | Deposit date: | 2009-08-18 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of human p70 ribosomal S6 kinase-1 regulation by activation loop phosphorylation.
J.Biol.Chem., 285, 2010
|
|
3BX7
 
 | |
3C86
 
 | | OpdA from agrobacterium radiobacter with bound product diethyl thiophosphate from crystal soaking with tetraethyl dithiopyrophosphate- 1.8 A | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, FE (II) ION, ... | | Authors: | Ollis, D.L, Jackson, C.J, Foo, J.L, Kim, H.K, Carr, P.D, Liu, J.W, Salem, G. | | Deposit date: | 2008-02-10 | | Release date: | 2008-02-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In crystallo capture of a Michaelis complex and product-binding modes of a bacterial phosphotriesterase
J.Mol.Biol., 375, 2008
|
|
3BX8
 
 | | Engineered Human Lipocalin 2 (LCN2), apo-form | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ENGINEERED HUMAN LIPOCALIN 2, PENTAETHYLENE GLYCOL | | Authors: | Schonfeld, D.L, Chatwell, L, Skerra, A. | | Deposit date: | 2008-01-11 | | Release date: | 2009-01-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High affinity molecular recognition and functional blockade of CTLA-4 by an engineered human lipocalin
To be Published
|
|
8GJV
 
 | |
4CAK
 
 | | Three-dimensional reconstruction of intact human integrin alphaIIbbeta3 in a phospholipid bilayer nanodisc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Integrin alpha-IIb, ... | | Authors: | Choi, W.S, Rice, W.J, Stokes, D.L, Coller, B.S. | | Deposit date: | 2013-10-08 | | Release date: | 2013-10-30 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (20.5 Å) | | Cite: | Three-Dimensional Reconstruction of Intact Human Integrin Alphaiibbeta3; New Implications for Activation-Dependent Ligand Binding.
Blood, 122, 2013
|
|
4EW1
 
 | | High resolution structure of human glycinamide ribonucleotide transformylase in apo form. | | Descriptor: | PHOSPHATE ION, SULFATE ION, Trifunctional purine biosynthetic protein adenosine-3 | | Authors: | Connelly, S, DeMartino, K, Boger, D.L, Wilson, I.A. | | Deposit date: | 2012-04-26 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.522 Å) | | Cite: | Biological and Structural Evaluation of 10R- and 10S-Methylthio-DDACTHF Reveals a New Role for Sulfur in Inhibition of Glycinamide Ribonucleotide Transformylase.
Biochemistry, 52, 2013
|
|
4EW3
 
 | | The structure of human glycinamide ribonucleotide transformylase in complex with 10R-methylthio-DDATHF. | | Descriptor: | N-({4-[(1R)-4-(2,4-diamino-6-oxo-1,6-dihydropyrimidin-5-yl)-1-(methylsulfanyl)butyl]phenyl}carbonyl)-L-glutamic acid, PHOSPHATE ION, SULFATE ION, ... | | Authors: | Connelly, S, DeMartino, K, Boger, D.L, Wilson, I.A. | | Deposit date: | 2012-04-26 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Biological and Structural Evaluation of 10R- and 10S-Methylthio-DDACTHF Reveals a New Role for Sulfur in Inhibition of Glycinamide Ribonucleotide Transformylase.
Biochemistry, 52, 2013
|
|
8OHN
 
 | | Human Coronavirus HKU1 spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Pronker, M.F, Hurdiss, D.L. | | Deposit date: | 2023-03-21 | | Release date: | 2023-08-02 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Sialoglycan binding triggers spike opening in a human coronavirus.
Nature, 624, 2023
|
|
