8UW0
 
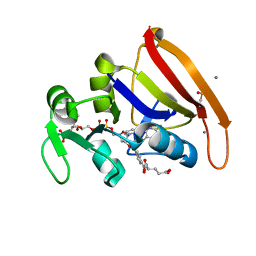 | |
8UVZ
 
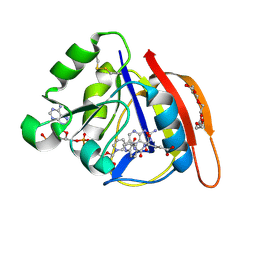 | |
7JMT
 
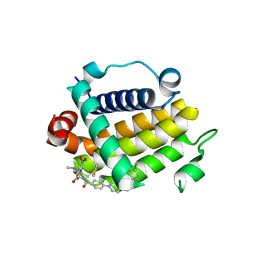 | | Crystal structure of schistosome BCL-2 bound to ABT-737 | | Descriptor: | 4-{4-[(4'-CHLOROBIPHENYL-2-YL)METHYL]PIPERAZIN-1-YL}-N-{[4-({(1R)-3-(DIMETHYLAMINO)-1-[(PHENYLTHIO)METHYL]PROPYL}AMINO)-3-NITROPHENYL]SULFONYL}BENZAMIDE, BCL-2 protein | | Authors: | Smith, N.A, Smith, B.J, Lee, E.F, Colman, P.M, Fairlie, W.D. | | Deposit date: | 2020-08-02 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Optimization of Benzothiazole and Thiazole Hydrazones as Inhibitors of Schistosome BCL-2.
Acs Infect Dis., 7, 2021
|
|
1R7S
 
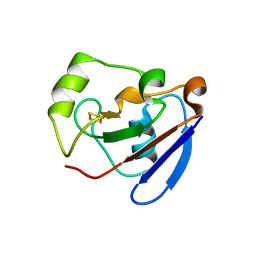 | | PUTIDAREDOXIN (Fe2S2 ferredoxin), C73G mutant | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Putidaredoxin | | Authors: | Smith, N, Mayhew, M, Kelly, H, Robinson, H, Heroux, A, Holden, M.J, Gallagher, D.T. | | Deposit date: | 2003-10-22 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of C73G putidaredoxin from Pseudomonas putida.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
4UX6
 
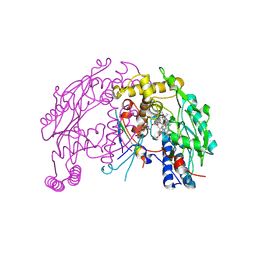 | | The discovery of novel, potent and highly selective inhibitors of inducible nitric oxide synthase (iNOS) | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, NITRIC OXIDE SYNTHASE, INDUCIBLE, ... | | Authors: | Cheshire, D.R, Andrews, G, Beaton, H.G, Birkinshaw, T, Boughton-Smith, N, Connolly, S, Cook, T.R, Cooper, A, Cooper, S.L, Cox, D, Dixon, J, Gensmantel, N, Hamley, P.J, Harrison, R, Hartopp, P, Kack, H, Luker, T, Mete, A, Millichip, I, Nicholls, D.J, Pimm, A.D, St-Gallay, S.A, Wallace, A.V. | | Deposit date: | 2014-08-19 | | Release date: | 2014-10-08 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Discovery of Novel, Potent and Highly Selective Inhibitors of Inducible Nitric Oxide Synthase (Inos).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
8TSX
 
 | |
8TT1
 
 | |
8TSU
 
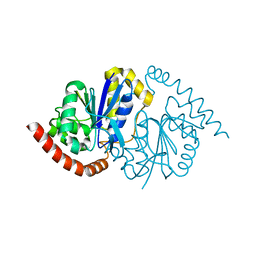 | |
8TT5
 
 | | Pseudomonas fluorescens isocyanide hydratase pH=8.3 | | Descriptor: | 1,2-ETHANEDIOL, Isonitrile hydratase InhA | | Authors: | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | Deposit date: | 2023-08-12 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Changes in an Enzyme Ensemble During Catalysis Observed by High Resolution XFEL Crystallography.
Biorxiv, 2023
|
|
8TSZ
 
 | |
8TT0
 
 | |
8TT4
 
 | | Pseudomonas fluorescens isocyanide hydratase pH=6.0 | | Descriptor: | 1,2-ETHANEDIOL, Isonitrile hydratase InhA | | Authors: | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | Deposit date: | 2023-08-12 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Changes in an Enzyme Ensemble During Catalysis Observed by High Resolution XFEL Crystallography.
Biorxiv, 2023
|
|
8TSY
 
 | |
8TT2
 
 | | Pseudomonas fluorescens isocyanide hydratase pH=5.4 | | Descriptor: | 1,2-ETHANEDIOL, Isonitrile hydratase InhA | | Authors: | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | Deposit date: | 2023-08-12 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Changes in an Enzyme Ensemble During Catalysis Observed by High Resolution XFEL Crystallography.
Biorxiv, 2023
|
|
8VQ1
 
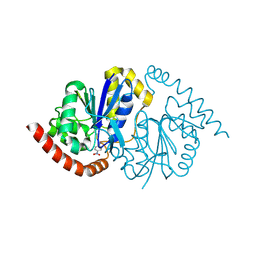 | | Pseudomonas fluorescens G150T isocyanide hydratase at 298 K XFEL data, thioimidate intermediate | | Descriptor: | CHLORIDE ION, Isonitrile hydratase InhA, N-(4-nitrophenyl)methanimine | | Authors: | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | Deposit date: | 2024-01-17 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Changes in an enzyme ensemble during catalysis observed by high-resolution XFEL crystallography.
Sci Adv, 10, 2024
|
|
8VPW
 
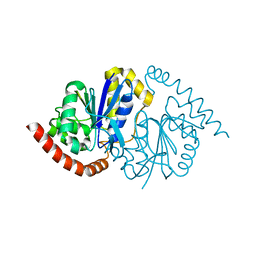 | | Pseudomonas fluorescens G150T isocyanide hydratase at 298 K XFEL data, free enzyme | | Descriptor: | CHLORIDE ION, Isonitrile hydratase InhA | | Authors: | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | Deposit date: | 2024-01-17 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Changes in an enzyme ensemble during catalysis observed by high-resolution XFEL crystallography.
Sci Adv, 10, 2024
|
|
6X44
 
 | | High Resolution Crystal Structure Analysis of SERA5 proenzyme from plasmodium falciparum | | Descriptor: | DI(HYDROXYETHYL)ETHER, Serine repeat antigen 5 | | Authors: | Clarke, O.B, Smith, N.A, Lee, M, Smith, B.J. | | Deposit date: | 2020-05-22 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19733953 Å) | | Cite: | Structure of the Plasmodium falciparum PfSERA5 pseudo-zymogen.
Protein Sci., 29, 2020
|
|
6X42
 
 | | High Resolution Crystal Structure Analysis of SERA5E from plasmodium falciparum | | Descriptor: | 1,2-ETHANEDIOL, 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, CALCIUM ION, ... | | Authors: | Clarke, O.B, Smith, N.A, Lee, M, Smith, B.J. | | Deposit date: | 2020-05-21 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of the Plasmodium falciparum PfSERA5 pseudo-zymogen.
Protein Sci., 29, 2020
|
|
6DCN
 
 | | Bcl-xL complex with Beclin 1 BH3 domain T108pThr | | Descriptor: | BCL-xl protein, Beclin-1 | | Authors: | Lee, E.F, Smith, B.J, Smith, N.A, Yao, S, Fairlie, W.D. | | Deposit date: | 2018-05-07 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.444 Å) | | Cite: | Structural insights into BCL2 pro-survival protein interactions with the key autophagy regulator BECN1 following phosphorylation by STK4/MST1.
Autophagy, 15, 2019
|
|
6DCO
 
 | | Bcl-xL complex with Beclin 1 BH3 domain T108D | | Descriptor: | Bcl-2-like protein 1,Bcl-2-like protein 1, Beclin-1, CHLORIDE ION | | Authors: | Lee, E.F, Smith, B.J, Smith, N.A, Yao, S, Fairlie, W.D. | | Deposit date: | 2018-05-07 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Structural insights into BCL2 pro-survival protein interactions with the key autophagy regulator BECN1 following phosphorylation by STK4/MST1.
Autophagy, 15, 2019
|
|
1OMO
 
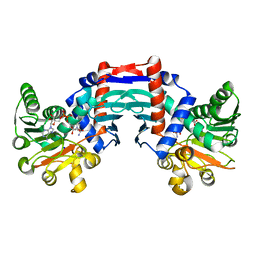 | | alanine dehydrogenase dimer w/bound NAD (archaeal) | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, alanine dehydrogenase | | Authors: | Gallagher, D.T, Smith, N.N, Holden, M.J, Schroeder, I, Monbouquette, H.G. | | Deposit date: | 2003-02-25 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure of alanine dehydrogenase from Archaeoglobus: active site analysis and relation to bacterial cyclodeaminases and mammalian mu crystallin.
J.Mol.Biol., 342, 2004
|
|
2AHC
 
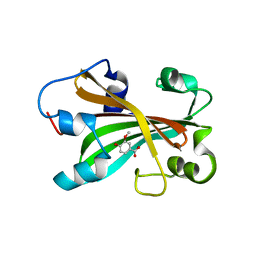 | | Chorismate lyase with inhibitor Vanilate | | Descriptor: | 4-HYDROXY-3-METHOXYBENZOATE, Chorismate lyase | | Authors: | Gallagher, D.T, Smith, N.N. | | Deposit date: | 2005-07-27 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analysis of ligand binding and catalysis in chorismate lyase
Arch.Biochem.Biophys., 445, 2006
|
|
2C8N
 
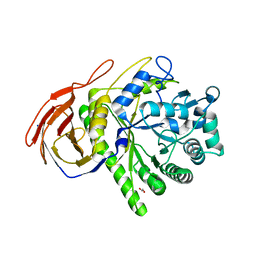 | | The Structure of a family 51 arabinofuranosidase, Araf51, from Clostridium thermocellum in complex with 1,3-linked arabinoside of xylobiose. | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-L-ARABINOFURANOSIDASE, alpha-L-arabinofuranose-(1-3)-alpha-D-xylopyranose | | Authors: | Taylor, E.J, Smith, N.L, Turkenburg, J.P, D'Souza, S, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2005-12-06 | | Release date: | 2005-12-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Insight Into the Ligand Specificity of a Thermostable Family 51 Arabinofuranosidase, Araf51, from Clostridium Thermocellum.
Biochem.J., 395, 2006
|
|
2C7F
 
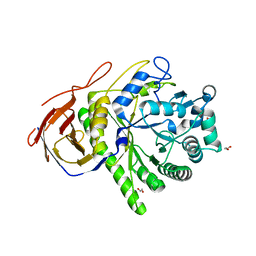 | | The Structure of a family 51 arabinofuranosidase, Araf51, from Clostridium thermocellum in complex with 1,5-alpha-L-Arabinotriose. | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-L-ARABINOFURANOSIDASE, alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose, ... | | Authors: | Taylor, E.J, Smith, N.L, Turkenburg, J.P, D'Souza, S, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2005-11-23 | | Release date: | 2005-12-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insight Into the Ligand Specificity of a Thermostable Family 51 Arabinofuranosidase, Araf51, from Clostridium Thermocellum.
Biochem.J., 395, 2006
|
|
2C3F
 
 | | The structure of a group A streptococcal phage-encoded tail-fibre showing hyaluronan lyase activity. | | Descriptor: | HYALURONIDASE, PHAGE ASSOCIATED, SODIUM ION | | Authors: | Taylor, E.J, Smith, N.L, Linsay, A.-M, Charnock, S.J, Turkenburg, J.P, Dodson, E.J, Davies, G.J, Black, G.W. | | Deposit date: | 2005-10-06 | | Release date: | 2005-11-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure of a Group a Streptococcal Phage-Encoded Virulence Factor Reveals Catalytically Active Triple-Stranded Beta-Helix
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
