6R2D
 
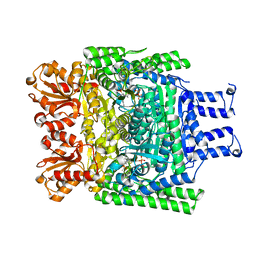 | | Crystal structure of the SucA domain of Mycobacterium smegmatis KGD after soaking with succinylphosphonate phosphonoethyl ester, followed by temperature increase | | Descriptor: | (4~{S})-4-[(2~{R})-3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-5-[2-[oxidanyl(phosphonooxy)phosphoryl]oxyethyl]-2~{H}-1,3-thiazol-2-yl]-4-[ethoxy(oxidanyl)phosphoryl]-4-oxidanyl-butanoic acid, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2019-03-15 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational transitions in the active site of mycobacterial 2-oxoglutarate dehydrogenase upon binding phosphonate analogues of 2-oxoglutarate: From a Michaelis-like complex to ThDP adducts.
J.Struct.Biol., 208, 2019
|
|
6R2C
 
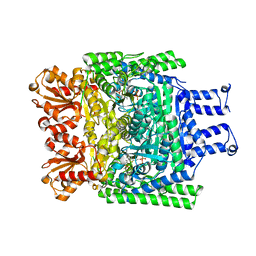 | | Crystal structure of the SucA domain of Mycobacterium smegmatis KGD after soaking with succinylphosphonate phosphonoethyl ester (PESP) | | Descriptor: | 4-[ethoxy(oxidanyl)phosphoryl]-4-oxidanylidene-butanoic acid, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2019-03-15 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Conformational transitions in the active site of mycobacterial 2-oxoglutarate dehydrogenase upon binding phosphonate analogues of 2-oxoglutarate: From a Michaelis-like complex to ThDP adducts.
J.Struct.Biol., 208, 2019
|
|
6R2B
 
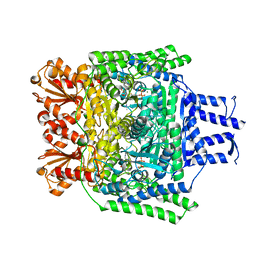 | | Crystal structure of the SucA domain of Mycobacterium smegmatis KGD after soaking with succinylphosphonate | | Descriptor: | (4~{S})-4-[(2~{R})-3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-5-[2-[oxidanyl(phosphonooxy)phosphoryl]oxyethyl]-2~{H}-1,3-thiazol-2-yl]-4-oxidanyl-4-phosphono-butanoic acid, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2019-03-15 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Conformational transitions in the active site of mycobacterial 2-oxoglutarate dehydrogenase upon binding phosphonate analogues of 2-oxoglutarate: From a Michaelis-like complex to ThDP adducts.
J.Struct.Biol., 208, 2019
|
|
6R29
 
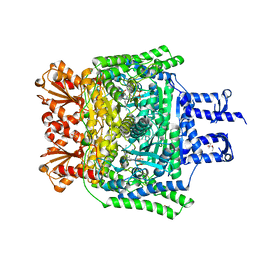 | | Crystal structure of the SucA domain of Mycobacterium smegmatis KGD cocrystallized with succinylphosphonate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (4~{S})-4-[(2~{R})-3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-5-[2-[oxidanyl(phosphonooxy)phosphoryl]oxyethyl]-2~{H}-1,3-thiazol-2-yl]-4-oxidanyl-4-phosphono-butanoic acid, MAGNESIUM ION, ... | | Authors: | Wagner, T, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2019-03-15 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Conformational transitions in the active site of mycobacterial 2-oxoglutarate dehydrogenase upon binding phosphonate analogues of 2-oxoglutarate: From a Michaelis-like complex to ThDP adducts.
J.Struct.Biol., 208, 2019
|
|
6ZZN
 
 | |
6ZZJ
 
 | | Crystal structure of the catalytic domain of Corynebacterium glutamicum acetyltransferase AceF (E2p) in complex with oxidized CoA. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, OXIDIZED COENZYME A | | Authors: | Bruch, E.M, Lexa-Sapart, N, Bellinzoni, M. | | Deposit date: | 2020-08-04 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Actinobacteria challenge the paradigm: A unique protein architecture for a well-known, central metabolic complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6ZZI
 
 | |
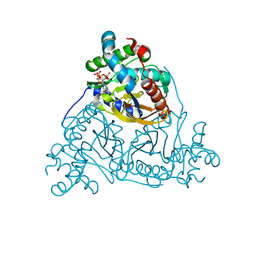
6ZZK
 
 | |
6ZZM
 
 | |
6ZZL
 
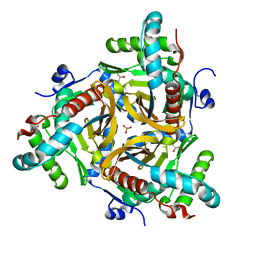 | |
4N9W
 
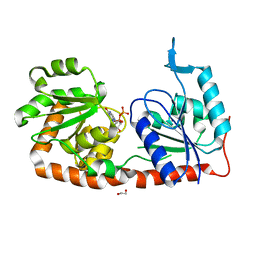 | | Crystal structure of phosphatidyl mannosyltransferase PimA | | Descriptor: | 1,2-ETHANEDIOL, GDP-mannose-dependent alpha-(1-2)-phosphatidylinositol mannosyltransferase, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Giganti, D, Albesa-Jove, D, Bellinzoni, M, Guerin, M.E, Alzari, P.M. | | Deposit date: | 2013-10-21 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Secondary structure reshuffling modulates glycosyltransferase function at the membrane.
Nat.Chem.Biol., 11, 2015
|
|
4NC9
 
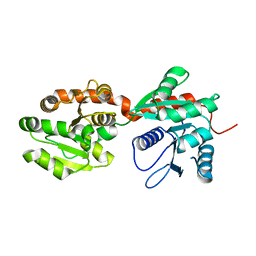 | | Crystal structure of phosphatidyl mannosyltransferase PimA | | Descriptor: | GDP-mannose-dependent alpha-(1-2)-phosphatidylinositol mannosyltransferase | | Authors: | Giganti, D, Albesa-Jove, D, Bellinzoni, M, Guerin, M.E, Alzari, P.M. | | Deposit date: | 2013-10-24 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.192 Å) | | Cite: | Secondary structure reshuffling modulates glycosyltransferase function at the membrane.
Nat.Chem.Biol., 11, 2015
|
|
2V7S
 
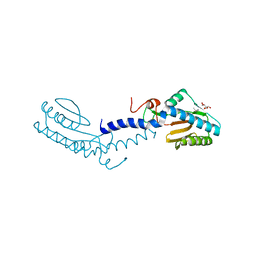 | | Crystal structure of the putative lipoprotein LppA from Mycobacterium tuberculosis | | Descriptor: | GLYCEROL, PROBABLE CONSERVED LIPOPROTEIN LPPA | | Authors: | Grana, M, Miras, I, Haouz, A, Winter, N, Buschiazzo, A, Bellinzoni, M, Alzari, P.M. | | Deposit date: | 2007-08-01 | | Release date: | 2008-08-26 | | Last modified: | 2011-09-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structure of Mycobacterium Tuberculosis Lppa, a Lipoprotein Confined to Pathogenic Mycobacteria.
Proteins, 78, 2010
|
|
2Y0P
 
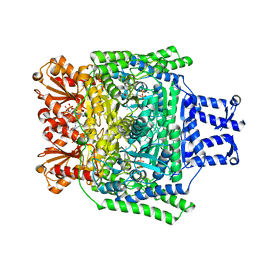 | | Crystal structure of the SucA domain of Mycobacterium smegmatis alpha- ketoglutarate decarboxylase in complex with the enamine-ThDP intermediate and acetyl-CoA | | Descriptor: | (4E)-4-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thiazol-2(3H)-ylidene}-4-hydroxybutanoic acid, 2-OXOGLUTARATE DECARBOXYLASE, ACETYL COENZYME *A, ... | | Authors: | Wagner, T, Bellinzoni, M, Wehenkel, A.M, O'Hare, H.M, Alzari, P.M. | | Deposit date: | 2010-12-07 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional Plasticity and Allosteric Regulation of Alpha-Ketoglutarate Decarboxylase in Central Mycobacterial Metabolism.
Chem.Biol., 18, 2011
|
|
2XTA
 
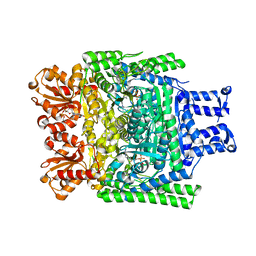 | | Crystal structure of the SucA domain of Mycobacterium smegmatis alpha- ketoglutarate decarboxylase in complex with acetyl-CoA (triclinic form) | | Descriptor: | 2-OXOGLUTARATE DECARBOXYLASE, ACETYL COENZYME *A, CALCIUM ION, ... | | Authors: | Wagner, T, Bellinzoni, M, Wehenkel, A.M, O'Hare, H.M, Alzari, P.M. | | Deposit date: | 2010-10-05 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional Plasticity and Allosteric Regulation of Alpha-Ketoglutarate Decarboxylase in Central Mycobacterial Metabolism.
Chem.Biol., 18, 2011
|
|
2YID
 
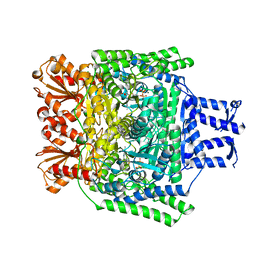 | | Crystal structure of the SucA domain of Mycobacterium smegmatis alpha- ketoglutarate decarboxylase in complex with the enamine-ThDP intermediate | | Descriptor: | (4E)-4-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thiazol-2(3H)-ylidene}-4-hydroxybutanoic acid, 2-OXOGLUTARATE DECARBOXYLASE, CALCIUM ION, ... | | Authors: | Wagner, T, Bellinzoni, M, Wehenkel, A.M, O'Hare, H.M, Alzari, P.M. | | Deposit date: | 2011-05-11 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Functional Plasticity and Allosteric Regulation of Alpha-Ketoglutarate Decarboxylase in Central Mycobacterial Metabolism.
Chem.Biol., 18, 2011
|
|
2YIC
 
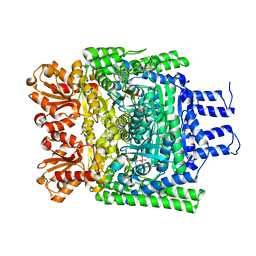 | | Crystal structure of the SucA domain of Mycobacterium smegmatis alpha- ketoglutarate decarboxylase (triclinic form) | | Descriptor: | 2-OXOGLUTARATE DECARBOXYLASE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Bellinzoni, M, Wehenkel, A.M, O'Hare, H.M, Alzari, P.M. | | Deposit date: | 2011-05-11 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Functional Plasticity and Allosteric Regulation of Alpha-Ketoglutarate Decarboxylase in Central Mycobacterial Metabolism.
Chem.Biol., 18, 2011
|
|
2XT6
 
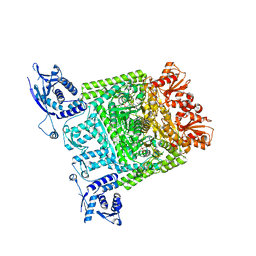 | | Crystal structure of Mycobacterium smegmatis alpha-ketoglutarate decarboxylase homodimer (orthorhombic form) | | Descriptor: | 2-OXOGLUTARATE DECARBOXYLASE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Bellinzoni, M, Wehenkel, A.M, O'Hare, H.M, Alzari, P.M. | | Deposit date: | 2010-10-05 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Functional Plasticity and Allosteric Regulation of Alpha-Ketoglutarate Decarboxylase in Central Mycobacterial Metabolism.
Chem.Biol., 18, 2011
|
|
2V06
 
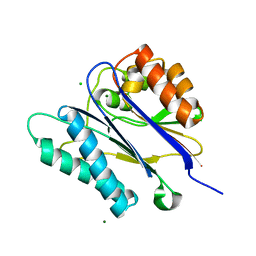 | | Crystal structure of the PPM Ser-Thr phosphatase MsPP from Mycobacterium smegmatis at pH 5.5 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, SER-THR PHOSPHATASE MSPP | | Authors: | Wehenkel, A, Bellinzoni, M, Schaeffer, F, Villarino, A, Alzari, P.M. | | Deposit date: | 2007-05-08 | | Release date: | 2007-10-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural and Binding Studies of the Three-Metal Center in Two Mycobacterial Ppm Ser/Thr Protein Phosphatases.
J.Mol.Biol., 374, 2007
|
|
3ZHR
 
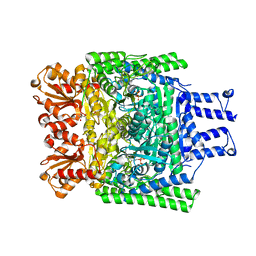 | | Crystal structure of the H747A mutant of the SucA domain of Mycobacterium smegmatis KGD showing the active site lid closed | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Barilone, N, Bellinzoni, M, Alzari, P.M. | | Deposit date: | 2012-12-24 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Dual Conformation of the Post-Decarboxylation Intermediate is Associated with Distinct Enzyme States in Mycobacterial Alpha-Ketoglutarate Decarboxylase (Kgd).
Biochem.J., 457, 2014
|
|
3ZHV
 
 | | Crystal structure of the SucA domain of Mycobacterium smegmatis KGD, post-decarboxylation intermediate from pyruvate (2-hydroxyethyl-ThDP) | | Descriptor: | 2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-2-[(1S)-1-oxidanylethyl]-1,3-thiazol-3-ium-5-yl]ethyl phosphono hydrogen phosphate, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Barilone, N, Bellinzoni, M, Alzari, P.M. | | Deposit date: | 2012-12-24 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Dual Conformation of the Post-Decarboxylation Intermediate is Associated with Distinct Enzyme States in Mycobacterial Alpha-Ketoglutarate Decarboxylase (Kgd).
Biochem.J., 457, 2014
|
|
3ZHT
 
 | | Crystal structure of the SucA domain of Mycobacterium smegmatis KGD, first post-decarboxylation intermediate from 2-oxoadipate | | Descriptor: | (5S)-5-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-4-methyl-5-(2-{[(phosphonatooxy)phosphinato]oxy}ethyl)-1,3-thiazol-3-ium-2-yl}-5-hydroxypentanoate, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Barilone, N, Bellinzoni, M, Alzari, P.M. | | Deposit date: | 2012-12-24 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Dual Conformation of the Post-Decarboxylation Intermediate is Associated with Distinct Enzyme States in Mycobacterial Alpha-Ketoglutarate Decarboxylase (Kgd).
Biochem.J., 457, 2014
|
|
3ZHQ
 
 | | Crystal structure of the H747A mutant of the SucA domain of Mycobacterium smegmatis KGD | | Descriptor: | CALCIUM ION, MAGNESIUM ION, MULTIFUNCTIONAL 2-OXOGLUTARATE METABOLISM ENZYME, ... | | Authors: | Wagner, T, Barilone, N, Bellinzoni, M, Alzari, P.M. | | Deposit date: | 2012-12-24 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Dual Conformation of the Post-Decarboxylation Intermediate is Associated with Distinct Enzyme States in Mycobacterial Alpha-Ketoglutarate Decarboxylase (Kgd).
Biochem.J., 457, 2014
|
|
3ZHS
 
 | | Crystal structure of the SucA domain of Mycobacterium smegmatis KGD, first post-decarboxylation intermediate from alpha-ketoglutarate | | Descriptor: | (4S)-4-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3lambda~5~-thiazol-2-yl}-4-hydroxybutanoic acid, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Barilone, N, Bellinzoni, M, Alzari, P.M. | | Deposit date: | 2012-12-24 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Dual Conformation of the Post-Decarboxylation Intermediate is Associated with Distinct Enzyme States in Mycobacterial Alpha-Ketoglutarate Decarboxylase (Kgd).
Biochem.J., 457, 2014
|
|
3ZHU
 
 | | Crystal structure of the SucA domain of Mycobacterium smegmatis KGD, second post-decarboxylation intermediate from 2-oxoadipate | | Descriptor: | (5R)-5-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-4-methyl-5-(2-{[(phosphonatooxy)phosphinato]oxy}ethyl)-1,3-thiazol-3-ium-2-yl}-5-hydroxypentanoate, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Barilone, N, Bellinzoni, M, Alzari, P.M. | | Deposit date: | 2012-12-24 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Dual Conformation of the Post-Decarboxylation Intermediate is Associated with Distinct Enzyme States in Mycobacterial Alpha-Ketoglutarate Decarboxylase (Kgd).
Biochem.J., 457, 2014
|
|
