3KS8
 
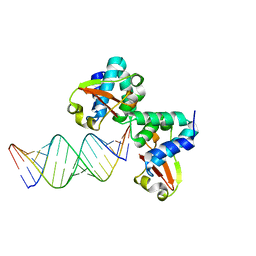 | | Crystal structure of Reston ebolavirus VP35 RNA binding domain in complex with 18bp dsRNA | | Descriptor: | 5'-R(*AP*GP*AP*AP*GP*GP*AP*GP*GP*GP*AP*GP*GP*GP*AP*GP*GP*A)-3', 5'-R(*UP*CP*CP*UP*CP*CP*CP*UP*CP*CP*CP*UP*CP*CP*UP*UP*CP*U)-3', Polymerase cofactor VP35 | | Authors: | Kimberlin, C.R, Bornholdt, Z.A, Li, S, Woods, V.L, Macrae, I.J, Saphire, E.O. | | Deposit date: | 2009-11-20 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression.
Proc.Natl.Acad.Sci.USA, 107, 2009
|
|
1ZSQ
 
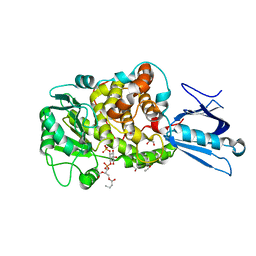 | | Crystal Structure of MTMR2 in complex with phosphatidylinositol 3-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-(BUTANOYLOXY)-1-{[(HYDROXY{[2,3,4,6-TETRAHYDROXY-5-(PHOSPHONOOXY)CYCLOHEXYL]OXY}PHOSPHORYL)OXY]METHYL}ETHYL BUTANOATE, Myotubularin-related protein 2 | | Authors: | Begley, M.J, Taylor, G.S, Brock, M.A, Ghosh, P, Woods, V.L, Dixon, J.E. | | Deposit date: | 2005-05-25 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Molecular basis for substrate recognition by MTMR2, a myotubularin family phosphoinositide phosphatase
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1ZVR
 
 | | Crystal Structure of MTMR2 in complex with phosphatidylinositol 3,5-bisphosphate | | Descriptor: | (1S)-2-(1-HYDROXYBUTOXY)-1-{[(HYDROXY{[(2R,3S,5R,6S)-2,4,6-TRIHYDROXY-3,5-BIS(PHOSPHONOOXY)CYCLOHEXYL]OXY}PHOSPHORYL)OXY]METHYL}ETHYL BUTYRATE, 1,2-ETHANEDIOL, Myotubularin-related protein 2 | | Authors: | Begley, M.J, Taylor, G.S, Brock, M.A, Ghosh, P, Woods, V.L, Dixon, J.E. | | Deposit date: | 2005-06-02 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Molecular basis for substrate recognition by MTMR2, a myotubularin family phosphoinositide phosphatase
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4F7Z
 
 | | Conformational dynamics of exchange protein directly activated by cAMP | | Descriptor: | GLYCEROL, Rap guanine nucleotide exchange factor 4 | | Authors: | White, M.A, Tsalkova, T.N, Mei, F.C, Liu, T, Woods, V.L, Cheng, X. | | Deposit date: | 2012-05-16 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analyses of a constitutively active mutant of exchange protein directly activated by cAMP.
Plos One, 7, 2012
|
|
3T5N
 
 | | 1.8A crystal structure of Lassa virus nucleoprotein in complex with ssRNA | | Descriptor: | NICKEL (II) ION, Nucleoprotein, RNA (5'-R(P*UP*AP*UP*CP*UP*C)-3') | | Authors: | Hastie, K.M, Liu, T, King, L.B, Ngo, N, Zandonatti, M.A, Woods, V.L, de la Torre, J.C, Saphire, E.O. | | Deposit date: | 2011-07-27 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.787 Å) | | Cite: | Crystal structure of the Lassa virus nucleoprotein-RNA complex reveals a gating mechanism for RNA binding.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3T5Q
 
 | | 3A structure of Lassa virus nucleoprotein in complex with ssRNA | | Descriptor: | Nucleoprotein, PHOSPHATE ION, RNA (5'-R(P*UP*AP*UP*CP*UP*C)-3'), ... | | Authors: | Hastie, K.M, Liu, T, King, L.B, Ngo, N, Zandonatti, M.A, Woods, V.L, de la Torre, J.C, Saphire, E.O. | | Deposit date: | 2011-07-27 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the Lassa virus nucleoprotein-RNA complex reveals a gating mechanism for RNA binding.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3KS4
 
 | | Crystal structure of Reston ebolavirus VP35 RNA binding domain | | Descriptor: | Polymerase cofactor VP35 | | Authors: | Kimberlin, C.R, Bornholdt, Z.A, Li, S, Woods, V.L, Macrae, I.J, Saphire, E.O. | | Deposit date: | 2009-11-20 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression.
Proc.Natl.Acad.Sci.USA, 107, 2009
|
|
3UAS
 
 | | Cytochrome P450 2B4 covalently bound to the mechanism-based inactivator 9-ethynylphenanthrene | | Descriptor: | 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Gay, S.C, Zhang, H, Shah, M.B, Stout, C.D, Halpert, J.R, Hollenberg, P.F. | | Deposit date: | 2011-10-21 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.939 Å) | | Cite: | Potent Mechanism-Based Inactivation of Cytochrome P450 2B4 by 9-Ethynylphenanthrene: Implications for Allosteric Modulation of Cytochrome P450 Catalysis.
Biochemistry, 52, 2013
|
|
1VJL
 
 | |
3TK3
 
 | | Cytochrome P450 2B4 mutant L437A in complex with 4-(4-chlorophenyl)imidazole | | Descriptor: | 4-(4-CHLOROPHENYL)IMIDAZOLE, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gay, S.C, Jang, H.H, Wilderman, P.R, Zhang, Q, Stout, C.D, Halpert, J.R. | | Deposit date: | 2011-08-25 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8001 Å) | | Cite: | Investigation by site-directed mutagenesis of the role of cytochrome P450 2B4 non-active-site residues in protein-ligand interactions based on crystal structures of the ligand-bound enzyme.
Febs J., 279, 2012
|
|
5I72
 
 | | Crystal structure of the oligomeric form of the Lassa virus matrix protein Z | | Descriptor: | RING finger protein Z, ZINC ION | | Authors: | Hastie, K, Zandonatti, M, Liu, T, Li, S, Woods Jr, V, Saphire, E.O. | | Deposit date: | 2016-02-16 | | Release date: | 2016-03-09 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Oligomeric Form of Lassa Virus Matrix Protein Z.
J.Virol., 90, 2016
|
|
3R1B
 
 | | Open crystal structure of cytochrome P450 2B4 covalently bound to the mechanism-based inactivator tert-butylphenylacetylene | | Descriptor: | (4-tert-butylphenyl)acetaldehyde, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B4, ... | | Authors: | Gay, S.C, Zhang, H, Stout, C.D, Hollenberg, P.F, Halpert, J.R. | | Deposit date: | 2011-03-09 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Analysis of Mammalian Cytochrome P450 2B4 Covalently Bound to the Mechanism-Based Inactivator tert-Butylphenylacetylene: Insight into Partial Enzymatic Activity.
Biochemistry, 50, 2011
|
|
3R1A
 
 | | Closed crystal structure of cytochrome P450 2B4 covalently bound to the mechanism-based inactivator tert-butylphenylacetylene | | Descriptor: | (4-tert-butylphenyl)acetaldehyde, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gay, S.C, Zhang, H, Stout, C.D, Hollenberg, P.F, Halpert, J.R. | | Deposit date: | 2011-03-09 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Analysis of Mammalian Cytochrome P450 2B4 Covalently Bound to the Mechanism-Based Inactivator tert-Butylphenylacetylene: Insight into Partial Enzymatic Activity.
Biochemistry, 50, 2011
|
|
3LGS
 
 | |
3MVR
 
 | |
4CSG
 
 | |
4CSF
 
 | | Structural insights into Toscana virus RNA encapsidation | | Descriptor: | NUCLEOPROTEIN, RNA (5'-R(*UP*GP*UP*GP*UP*UP*UP*CP*UP)-3') | | Authors: | Olal, D, Daumke, O. | | Deposit date: | 2014-03-07 | | Release date: | 2014-04-09 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Structural Insights Into RNA Encapsidation and Helical Assembly of the Toscana Virus Nucleoprotein.
Nucleic Acids Res., 42, 2014
|
|
4K3V
 
 | | Structure of Staphylococcus aureus MntC | | Descriptor: | ABC superfamily ATP binding cassette transporter, binding protein, MANGANESE (II) ION | | Authors: | Parris, K.D, Mosyak, L. | | Deposit date: | 2013-04-11 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-Dimensional Structure and Biophysical Characterization of Staphylococcus aureus Cell Surface Antigen-Manganese Transporter MntC.
J.Mol.Biol., 425, 2013
|
|
3COC
 
 | |
3COD
 
 | | Crystal Structure of T90A/D115A mutant of Bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Joh, N.H, Min, A, Faham, S, Bowie, J.U. | | Deposit date: | 2008-03-27 | | Release date: | 2008-04-08 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Modest stabilization by most hydrogen-bonded side-chain interactions in membrane proteins.
Nature, 453, 2008
|
|
3VE0
 
 | | Crystal structure of Sudan Ebolavirus Glycoprotein (strain Boniface) bound to 16F6 | | Descriptor: | 16F6 Antibody chain A, 16F6 Antibody chain B, Envelope glycoprotein, ... | | Authors: | Saphire, E.O, Bale, S, Dias, J.M. | | Deposit date: | 2012-01-06 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.353 Å) | | Cite: | Structural basis for differential neutralization of ebolaviruses.
Viruses, 4, 2012
|
|
4RKK
 
 | | Structure of a product bound phosphatase | | Descriptor: | Laforin, PHOSPHATE ION, alpha-D-glucopyranose, ... | | Authors: | Vander Kooi, C.W. | | Deposit date: | 2014-10-13 | | Release date: | 2015-01-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural mechanism of laforin function in glycogen dephosphorylation and lafora disease.
Mol.Cell, 57, 2015
|
|
4HFW
 
 | | Anti Rotavirus Antibody | | Descriptor: | 6-26 Fab Heavy chain, 6-26 Fab Light chain, SULFATE ION | | Authors: | Spiller, B.W, Aiyegbo, M, Crowe, J.E. | | Deposit date: | 2012-10-05 | | Release date: | 2013-05-29 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Human Rotavirus VP6-Specific Antibodies Mediate Intracellular Neutralization by Binding to a Quaternary Structure in the Transcriptional Pore.
Plos One, 8, 2013
|
|
1O5L
 
 | |
3QAM
 
 | | Crystal Structure of Glu208Ala mutant of catalytic subunit of cAMP-dependent protein kinase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein kinase inhibitor, ... | | Authors: | Yang, J, Wu, J, Steichen, J, Taylor, S.S. | | Deposit date: | 2011-01-11 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | A conserved Glu-Arg salt bridge connects coevolved motifs that define the eukaryotic protein kinase fold.
J.Mol.Biol., 415, 2012
|
|
