1LSH
 
 | |
1YBA
 
 | | The active form of phosphoglycerate dehydrogenase | | Descriptor: | 2-OXOGLUTARIC ACID, D-3-phosphoglycerate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Thompson, J.R, Banaszak, L.J. | | Deposit date: | 2004-12-20 | | Release date: | 2005-04-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Vmax Regulation through Domain and Subunit Changes. The Active Form of Phosphoglycerate Dehydrogenase
Biochemistry, 44, 2005
|
|
1CBI
 
 | | APO-CELLULAR RETINOIC ACID BINDING PROTEIN I | | Descriptor: | CELLULAR RETINOIC ACID BINDING PROTEIN I | | Authors: | Thompson, J.R, Bratt, J.M, Banaszak, L.J. | | Deposit date: | 1995-07-12 | | Release date: | 1995-11-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of cellular retinoic acid binding protein I shows increased access to the binding cavity due to formation of an intermolecular beta-sheet.
J.Mol.Biol., 252, 1995
|
|
2GGM
 
 | | Human centrin 2 xeroderma pigmentosum group C protein complex | | Descriptor: | CALCIUM ION, Centrin-2, DNA-repair protein complementing XP-C cells | | Authors: | Thompson, J.R. | | Deposit date: | 2006-03-24 | | Release date: | 2006-04-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The structure of the human centrin 2-xeroderma pigmentosum group C protein complex.
J.Biol.Chem., 281, 2006
|
|
3DVI
 
 | |
4K07
 
 | | Crystal structure of the amyloid-forming immunoglobulin AL-103 cis-proline 95 mutant | | Descriptor: | Amyloidogenic immunoglobulin light chain protein AL-103, SULFATE ION | | Authors: | Thompson, J.R, Berkholz, D.S, Mahlum, E.W, Ramirez-Alvarado, M. | | Deposit date: | 2013-04-03 | | Release date: | 2013-10-30 | | Last modified: | 2014-01-22 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Kinetic control in protein folding for light chain amyloidosis and the differential effects of somatic mutations.
J.Mol.Biol., 426, 2014
|
|
2Q20
 
 | |
2Q1E
 
 | |
3CDC
 
 | |
3CDY
 
 | | AL-09 H87Y, immunoglobulin light chain variable domain | | Descriptor: | IMMUNOGLOBULIN LIGHT CHAIN | | Authors: | Baden, E.M, Randles, E.G, Aboagye, A.K, Thompson, J.R, Ramirez-Alvarado, M. | | Deposit date: | 2008-02-27 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural insights into the role of mutations in amyloidogenesis.
J.Biol.Chem., 283, 2008
|
|
3CDF
 
 | |
2ETX
 
 | | Crystal Structure of MDC1 Tandem BRCT Domains | | Descriptor: | Mediator of DNA damage checkpoint protein 1 | | Authors: | Wasielewski, E, Kim, Y, Joachimiak, A, Thompson, J.R, Mer, G. | | Deposit date: | 2005-10-27 | | Release date: | 2005-11-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Molecular Basis for the Association of Microcephalin (MCPH1) Protein with the Cell Division Cycle Protein 27 (Cdc27) Subunit of the Anaphase-promoting Complex.
J.Biol.Chem., 287, 2012
|
|
3U3Z
 
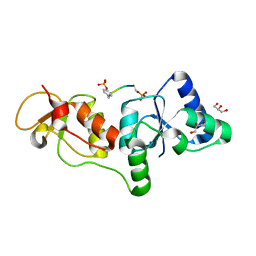 | | Structure of human microcephalin (MCPH1) tandem BRCT domains in complex with an H2A.X peptide phosphorylated at Ser139 and Tyr142 | | Descriptor: | GLYCEROL, Histone H2A.X peptide, Microcephalin | | Authors: | Singh, N, Thompson, J.R, Heroux, A, Mer, G. | | Deposit date: | 2011-10-06 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Dual recognition of phosphoserine and phosphotyrosine in histone variant H2A.X by DNA damage response protein MCPH1.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3U7A
 
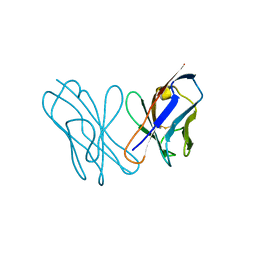 | | AL-09 Y32F Y96F | | Descriptor: | Amyloidogenic immunoglobulin light chain protein AL-09 Y32F Y96F, variable domain | | Authors: | DiCostanzo, A.C, Thompson, J.R, Ramirez-Alvarado, M. | | Deposit date: | 2011-10-13 | | Release date: | 2012-07-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Tyrosine Residues mediate crucial interactions in amyloid formation for immunoglobulin light chains
To be Published
|
|
3L1X
 
 | |
3KTF
 
 | |
3L1Z
 
 | |
3L1Y
 
 | |
4W8R
 
 | |
4W8S
 
 | |
4W8T
 
 | |
5KZ5
 
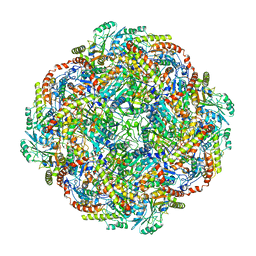 | | Architecture of the Human Mitochondrial Iron-Sulfur Cluster Assembly Machinery: the Complex Formed by the Iron Donor, the Sulfur Donor, and the Scaffold | | Descriptor: | Cysteine desulfurase, mitochondrial, Frataxin, ... | | Authors: | Gakh, O, Ranatunga, W, Smith, D.Y, Ahlgren, E.C, Al-Karadaghi, S, Thompson, J.R, Isaya, G. | | Deposit date: | 2016-07-22 | | Release date: | 2016-08-31 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (14.3 Å) | | Cite: | Architecture of the Human Mitochondrial Iron-Sulfur Cluster Assembly Machinery.
J.Biol.Chem., 291, 2016
|
|
2B02
 
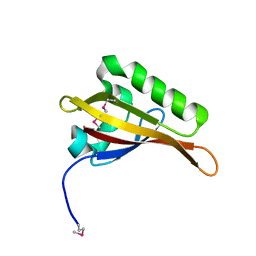 | | Crystal Structure of ARNT PAS-B Domain | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator | | Authors: | Lee, J, Botuyan, M.V, Nomine, Y, Ohh, M, Thompson, J.R, Mer, G. | | Deposit date: | 2005-09-12 | | Release date: | 2006-10-24 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure and Binding Properties of ARNT PAS-B Heterodimerization Domain
To be Published
|
|
2FHD
 
 | | Crystal structure of Crb2 tandem tudor domains | | Descriptor: | DNA repair protein rhp9/CRB2, PHOSPHATE ION | | Authors: | Lee, J, Botuyan, M.V, Thompson, J.R, Mer, G. | | Deposit date: | 2005-12-23 | | Release date: | 2007-01-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair.
Cell(Cambridge,Mass.), 127, 2006
|
|
3PA6
 
 | |
