7PQG
 
 | |
7Q1U
 
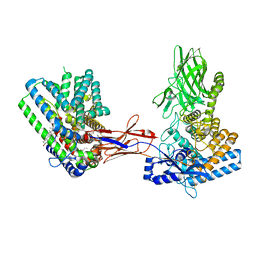 | | Structure of Hedgehog acyltransferase (HHAT) in complex with megabody 177 bound to non-hydrolysable palmitoyl-CoA (Composite Map) | | Descriptor: | CHOLESTEROL, MAGNESIUM ION, Megabody 177, ... | | Authors: | Coupland, C, Carrique, L, Siebold, C. | | Deposit date: | 2021-10-21 | | Release date: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure, mechanism, and inhibition of Hedgehog acyltransferase.
Mol.Cell, 81, 2021
|
|
6GS7
 
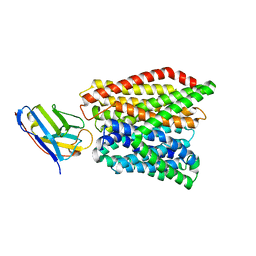 | | Crystal structure of peptide transporter DtpA-nanobody in glycine buffer | | Descriptor: | Dipeptide and tripeptide permease A, nanobody | | Authors: | Ural-Blimke, Y, Flayhan, A, Quistgaard, E.M, Loew, C. | | Deposit date: | 2018-06-13 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of Prototypic Peptide Transporter DtpA from E. coli in Complex with Valganciclovir Provides Insights into Drug Binding of Human PepT1.
J. Am. Chem. Soc., 141, 2019
|
|
5N7O
 
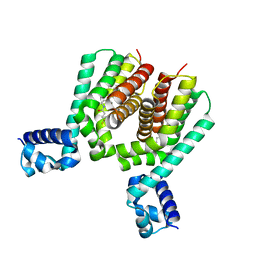 | | EthR2 in complex with SMARt-420 compound | | Descriptor: | 4,4,4-trifluoro-1-(3-phenyl-1-oxa-2,8-diazaspiro[4.5]dec-2-en-8-yl)butan-1-one, Probable transcriptional regulatory protein | | Authors: | Wohlkonig, A, Wintjens, R. | | Deposit date: | 2017-02-21 | | Release date: | 2017-04-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of the interaction between spiroisoxazoline SMARt-420 and the Mycobacterium tuberculosis repressor EthR2.
Biochem. Biophys. Res. Commun., 487, 2017
|
|
6QPG
 
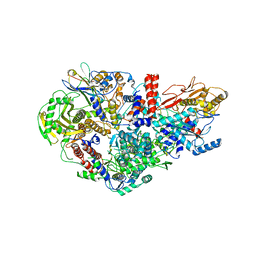 | | Influenza A virus Polymerase Heterotrimer A/nt/60/1968(H3N2) in complex with Nanobody NB8205 | | Descriptor: | Nanobody NB8205, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Fan, H.T, Keown, J.R, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-02-13 | | Release date: | 2019-09-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
6QXE
 
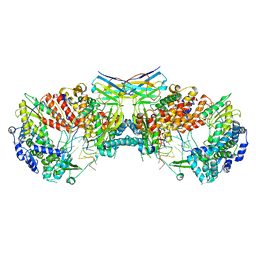 | | Influenza A virus (A/NT/60/1968) polymerase dimer of hetermotrimer in complex with 3'5' cRNA promoter and Nb8205 | | Descriptor: | Nb8205, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Carrique, L, Keown, J.R, Fan, H, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-04 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
6QX8
 
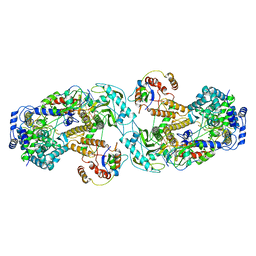 | | Influenza A virus (A/NT/60/1968) polymerase dimer of heterotrimer in complex with 5' cRNA promoter | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(P*AP*GP*CP*AP*AP*AP*AP*GP*CP*AP*GP*A)-3'), ... | | Authors: | Carrique, L, Keown, J.R, Fan, H, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-04 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
6HLU
 
 | |
6HER
 
 | | Mouse prion protein in complex with Nanobody 484 | | Descriptor: | GLYCEROL, Major prion protein, Nanobody 484, ... | | Authors: | Soror, S.H, Abskharon, R, Wohlkonig, A. | | Deposit date: | 2018-08-20 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | Structural evidence for the critical role of the prion protein hydrophobic region in forming an infectious prion.
Plos Pathog., 15, 2019
|
|
6HHD
 
 | | Mouse Prion Protein in complex with Nanobody 484 | | Descriptor: | CHLORIDE ION, GLYCEROL, Major prion protein, ... | | Authors: | Soror, S, Abskharon, R, Wohlkonig, A. | | Deposit date: | 2018-08-28 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structural evidence for the critical role of the prion protein hydrophobic region in forming an infectious prion.
Plos Pathog., 15, 2019
|
|
5IOF
 
 | |
2LAL
 
 | | CRYSTAL STRUCTURE DETERMINATION AND REFINEMENT AT 2.3 ANGSTROMS RESOLUTION OF THE LENTIL LECTIN | | Descriptor: | CALCIUM ION, LENTIL LECTIN (ALPHA CHAIN), LENTIL LECTIN (BETA CHAIN), ... | | Authors: | Loris, R, Steyaert, J, Maes, D, Lisgarten, J, Pickersgill, R, Wyns, L. | | Deposit date: | 1993-06-10 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of two crystal forms of lentil lectin at 1.8 A resolution.
Proteins, 20, 1994
|
|
3MKM
 
 | | Crystal structure of the E. coli pyrimidine nucleoside hydrolase YeiK (Apo-form) | | Descriptor: | CALCIUM ION, Putative uncharacterized protein YeiK | | Authors: | Garau, G, Fornili, A, Giabbai, B, Degano, M. | | Deposit date: | 2010-04-15 | | Release date: | 2010-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Energy Landscapes Associated with Macromolecular Conformational Changes from Endpoint Structures
J.Am.Chem.Soc., 132, 2010
|
|
5N1I
 
 | |
4DK3
 
 | | Structure of Editosome protein | | Descriptor: | RNA-editing complex protein MP81, single domain antibody VHH | | Authors: | Park, Y.-J, Hol, W. | | Deposit date: | 2012-02-03 | | Release date: | 2012-07-04 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | The structure of the C-terminal domain of the largest editosome interaction protein and its role in promoting RNA binding by RNA-editing ligase L2.
Nucleic Acids Res., 40, 2012
|
|
4DKA
 
 | | Structure of Editosome protein | | Descriptor: | RNA-editing complex protein MP81, SODIUM ION, single domain antibody VHH | | Authors: | Park, Y.-J, Hol, W. | | Deposit date: | 2012-02-03 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The structure of the C-terminal domain of the largest editosome interaction protein and its role in promoting RNA binding by RNA-editing ligase L2.
Nucleic Acids Res., 40, 2012
|
|
5N1C
 
 | |
4DK6
 
 | | Structure of Editosome protein | | Descriptor: | RNA-editing complex protein MP81, single domain antibody VHH | | Authors: | Park, Y.-J, Hol, W. | | Deposit date: | 2012-02-03 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The structure of the C-terminal domain of the largest editosome interaction protein and its role in promoting RNA binding by RNA-editing ligase L2.
Nucleic Acids Res., 40, 2012
|
|
5OCL
 
 | | Nanobody-anti-VGLUT nanobody complex | | Descriptor: | anti-llama nanobody, anti-vglut nanobody | | Authors: | Dutzler, R, Schenck, S, Kunz, L. | | Deposit date: | 2017-07-03 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Generation and Characterization of Anti-VGLUT Nanobodies Acting as Inhibitors of Transport.
Biochemistry, 56, 2017
|
|
1Q8F
 
 | |
6H02
 
 | |
4M3K
 
 | | Structure of a single domain camelid antibody fragment cAb-H7S in complex with the BlaP beta-lactamase from Bacillus licheniformis | | Descriptor: | Beta-lactamase, CHLORIDE ION, Camelid heavy-chain antibody variable fragment cAb-H7S | | Authors: | Pain, C, Kerff, F, Herman, R, Sauvage, E, Preumont, S, Charlier, P, Dumoulin, M. | | Deposit date: | 2013-08-06 | | Release date: | 2014-08-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Probing the mechanism of aggregation of polyQ model proteins with camelid heavy-chain antibody fragments
To be Published
|
|
4M3J
 
 | | Structure of a single-domain camelid antibody fragment cAb-H7S specific of the BlaP beta-lactamase from Bacillus licheniformis | | Descriptor: | Camelid heavy-chain antibody variable fragment cAb-H7S, SULFATE ION | | Authors: | Pain, C, Kerff, F, Herman, R, Sauvage, E, Preumont, S, Charlier, P, Dumoulin, M. | | Deposit date: | 2013-08-06 | | Release date: | 2014-08-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Probing the mechanism of aggregation of polyQ model proteins with camelid heavy-chain antibody fragments
To be Published
|
|
4N1H
 
 | | Structure of a single-domain camelid antibody fragment cAb-F11N in complex with the BlaP beta-lactamase from Bacillus licheniformis | | Descriptor: | Beta-lactamase, Camelid heavy-chain antibody variable fragment cAb-F11N | | Authors: | Pain, C, Kerff, F, Herman, R, Sauvage, E, Preumont, S, Charlier, P, Dumoulin, M. | | Deposit date: | 2013-10-04 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Probing the mechanism of aggregation of polyQ model proteins with camelid heavy-chain antibody fragments
To be Published
|
|
4GGN
 
 | | Malaria invasion machinery protein complex | | Descriptor: | Myosin A tail domain interacting protein MTIP, Myosin-A | | Authors: | Khamrui, S, Turley, S, Bergman, L.W, Hol, W.G.J. | | Deposit date: | 2012-08-06 | | Release date: | 2013-07-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | The structure of the D3 domain of Plasmodium falciparum myosin tail interacting protein MTIP in complex with a nanobody.
Mol.Biochem.Parasitol., 190, 2013
|
|
