7LNT
 
 | | Ternary complex of the Isopentenyl Phosphate Kinase from Candidatus methanomethylophilus alvus bound to benzyl monophosphate and ATP | | Descriptor: | (phenylmethyl) dihydrogen phosphate, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Thomas, L.M, Singh, S, Scull, E.M, Bourne, C.R. | | Deposit date: | 2021-02-08 | | Release date: | 2021-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular Basis for the Substrate Promiscuity of Isopentenyl Phosphate Kinase from Candidatus methanomethylophilus alvus .
Acs Chem.Biol., 17, 2022
|
|
7LNU
 
 | | Ternary complex of the Isopentenyl Phosphate Kinase from Candidatus methanomethylophilus alvus bound to isopentenyl monophosphate and ATP | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Thomas, L.M, Singh, S, Scull, E.M, Bourne, C.R. | | Deposit date: | 2021-02-08 | | Release date: | 2021-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Basis for the Substrate Promiscuity of Isopentenyl Phosphate Kinase from Candidatus methanomethylophilus alvus .
Acs Chem.Biol., 17, 2022
|
|
7LNV
 
 | | Apo Structure of Isopentenyl Phosphate Kinase from Candidatus methanomethylophilus alvus | | Descriptor: | GLYCEROL, Isopentenyl phosphate kinase, MALONATE ION | | Authors: | Thomas, L.M, Singh, S, Scull, E.M, Bourne, C.R. | | Deposit date: | 2021-02-08 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular Basis for the Substrate Promiscuity of Isopentenyl Phosphate Kinase from Candidatus methanomethylophilus alvus .
Acs Chem.Biol., 17, 2022
|
|
7SHF
 
 | | Cryo-EM structure of GPR158 coupled to the RGS7-Gbeta5 complex | | Descriptor: | (2S)-1-{[(S)-hydroxy{[(1s,2R,3R,4R,5S,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5E,8E,11E,14E)-icosa-5,8,11,14-tetraenoate, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHOLESTEROL, ... | | Authors: | Patil, D.N, Singh, S, Singh, A.K, Martemyanov, K.A. | | Deposit date: | 2021-10-08 | | Release date: | 2021-12-01 | | Last modified: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of human GPR158 receptor coupled to the RGS7-G beta 5 signaling complex.
Science, 375, 2022
|
|
7SHE
 
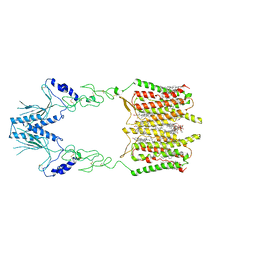 | | Cryo-EM structure of human GPR158 | | Descriptor: | (2S)-1-{[(S)-hydroxy{[(1s,2R,3R,4R,5S,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5E,8E,11E,14E)-icosa-5,8,11,14-tetraenoate, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHOLESTEROL, ... | | Authors: | Patil, D.N, Singh, S, Singh, A.K, Martemyanov, K.A. | | Deposit date: | 2021-10-08 | | Release date: | 2021-12-01 | | Last modified: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of human GPR158 receptor coupled to the RGS7-G beta 5 signaling complex.
Science, 375, 2022
|
|
5EYB
 
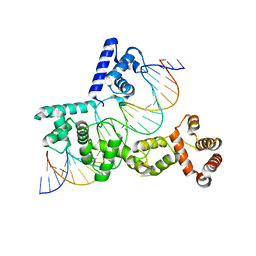 | | X-ray Structure of Reb1-Ter Complex | | Descriptor: | DNA (26-MER), DNA-binding protein reb1 | | Authors: | Jaiswal, R, Choudhury, M, Zaman, S, Singh, S, Santosh, V, Bastia, D, Escalante, C.R. | | Deposit date: | 2015-11-24 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Functional architecture of the Reb1-Ter complex of Schizosaccharomyces pombe.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7N9D
 
 | |
7S16
 
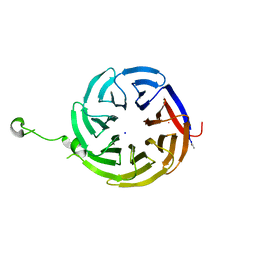 | | Crystal structure of alpha-COP-WD40 domain R57A mutant | | Descriptor: | Coatomer subunit alpha, SODIUM ION | | Authors: | Dey, D, Singh, S, Khan, S, Martin, M, Schnicker, N, Gakhar, L, Pierce, B, Hasan, S.S. | | Deposit date: | 2021-09-01 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | An extended motif in the SARS-CoV-2 spike modulates binding and release of host coatomer in retrograde trafficking
Commun Biol, 5, 2022
|
|
7S22
 
 | | Crystal structure of alpha-COP-WD40 domain | | Descriptor: | Coatomer subunit alpha | | Authors: | Dey, D, Singh, S, Khan, S, Martin, M, Schnicker, N, Gakhar, L, Pierce, B, Hasan, S.S. | | Deposit date: | 2021-09-02 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An extended motif in the SARS-CoV-2 spike modulates binding and release of host coatomer in retrograde trafficking
Commun Biol, 5, 2022
|
|
7S23
 
 | | Crystal structure of alpha-COP-WD40 domain, Y139A mutant | | Descriptor: | Coatomer subunit alpha | | Authors: | Dey, D, Singh, S, Khan, S, Martin, M, Schnicker, N, Gakhar, L, Pierce, B, Hasan, S.S. | | Deposit date: | 2021-09-03 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | An extended motif in the SARS-CoV-2 spike modulates binding and release of host coatomer in retrograde trafficking
Commun Biol, 5, 2022
|
|
3IAA
 
 | | Crystal Structure of CalG2, Calicheamicin Glycosyltransferase, TDP bound form | | Descriptor: | CalG2, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2009-07-13 | | Release date: | 2010-06-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.505 Å) | | Cite: | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3IA7
 
 | | Crystal Structure of CalG4, the Calicheamicin Glycosyltransferase | | Descriptor: | CALCIUM ION, CHLORIDE ION, CalG4 | | Authors: | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2009-07-13 | | Release date: | 2010-06-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4WS9
 
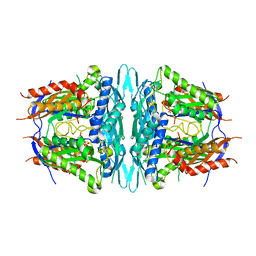 | | Crystal structure of sMAT N159G from Sulfolobus solfataricus | | Descriptor: | PHOSPHATE ION, S-adenosylmethionine synthase | | Authors: | Wang, F, Brady, E.L, Singh, S, Clinger, J.A, Huber, T.D, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2014-10-26 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Crystal structure of sMAT N159G from Sulfolobus solfataricus.
To Be Published
|
|
4XQ2
 
 | | Ensemble refinement of cystathione gamma lyase (CalE6) D7G from Micromonospora echinospora | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, CalE6, ... | | Authors: | Wang, F, Yennamalli, R.M, Singh, S, Tan, K, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-01-18 | | Release date: | 2015-04-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of cystathione gamma lyase (CalE6) from Micromonospora echinospora
To Be Published
|
|
4XR9
 
 | | Crystal structure of CalS8 from Micromonospora echinospora cocrystallized with NAD and TDP-glucose | | Descriptor: | CalS8, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Michalska, K, Bigelow, L, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R.M, Singh, S, Kharel, M.K, Thorson, J.S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-01-20 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of CalS8 from Micromonospora echinospora
To Be Published
|
|
7XNZ
 
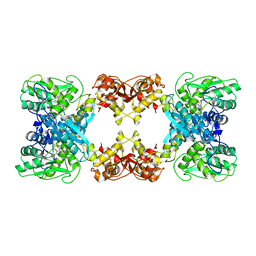 | | Native cystathionine beta-synthase of Mycobacterium tuberculosis. | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Putative cystathionine beta-synthase Rv1077 | | Authors: | Bandyopadhyay, P, Pramanick, I, Biswas, R, Sabarinath, P.S, Sreedharan, S, Singh, S, Rajmani, R, Laxman, S, Dutta, S, Singh, A. | | Deposit date: | 2022-04-30 | | Release date: | 2022-05-25 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | S-Adenosylmethionine-responsive cystathionine beta-synthase modulates sulfur metabolism and redox balance in Mycobacterium tuberculosis.
Sci Adv, 8, 2022
|
|
7XOH
 
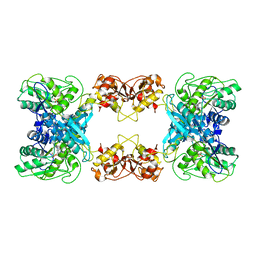 | | Cystathionine beta-synthase of Mycobacterium tuberculosis in the presence of S-adenosylmethionine. | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Putative cystathionine beta-synthase Rv1077 | | Authors: | Bandyopadhyay, P, Pramanick, I, Biswas, R, Sabarinath, P.S, Sreedharan, S, Singh, S, Rajmani, R, Laxman, S, Dutta, S, Singh, A. | | Deposit date: | 2022-05-01 | | Release date: | 2022-05-25 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | S-Adenosylmethionine-responsive cystathionine beta-synthase modulates sulfur metabolism and redox balance in Mycobacterium tuberculosis.
Sci Adv, 8, 2022
|
|
7XOY
 
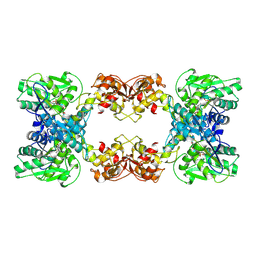 | | Cystathionine beta-synthase of Mycobacterium tuberculosis in the presence of S-adenosylmethionine and serine. | | Descriptor: | Putative cystathionine beta-synthase Rv1077, [3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-SERINE | | Authors: | Bandyopadhyay, P, Pramanick, I, Biswas, R, Sabarinath, P.S, Sreedharan, S, Singh, S, Rajmani, R, Laxman, S, Dutta, S, Singh, A. | | Deposit date: | 2022-05-01 | | Release date: | 2022-05-25 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | S-Adenosylmethionine-responsive cystathionine beta-synthase modulates sulfur metabolism and redox balance in Mycobacterium tuberculosis.
Sci Adv, 8, 2022
|
|
8GDY
 
 | | Crystal structure of the human PDI first domain with 9 mutations | | Descriptor: | 1,2-ETHANEDIOL, Protein disulfide-isomerase, THIOCYANATE ION | | Authors: | Forouhar, F, Banayan, N.E, Loughlin, B.L, Singh, S, Wong, V, Hunt, H.S, Handelman, S.K, Price, N, Hunt, J.F. | | Deposit date: | 2023-03-06 | | Release date: | 2024-01-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Systematic enhancement of protein crystallization efficiency by bulk lysine-to-arginine (KR) substitution.
Protein Sci., 33, 2024
|
|
8GDU
 
 | | Crystal structure of a mutant methyl transferase from Methanosarcina acetivorans, Northeast Structural Genomics Consortium (NESG) Target MvR53-11M | | Descriptor: | Methyltransferase domain-containing protein, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Forouhar, F, Banayan, N.E, Loughlin, B.L, Singh, S, Wong, V, Hunt, H.S, Handelman, S.K, Price, N, Hunt, J.F. | | Deposit date: | 2023-03-06 | | Release date: | 2024-01-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Systematic enhancement of protein crystallization efficiency by bulk lysine-to-arginine (KR) substitution.
Protein Sci., 33, 2024
|
|
5HOQ
 
 | | Apo structure of CalS11, TDP-rhamnose 3'-o-methyltransferase, an enzyme in Calicheamicin biosynthesis | | Descriptor: | SULFATE ION, TDP-rhamnose 3'-O-methyltransferase (CalS11) | | Authors: | Han, L, Helmich, K.E, Singh, S, Thorson, J.S, Bingman, C.A, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis | | Deposit date: | 2016-01-19 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | Loop dynamics of thymidine diphosphate-rhamnose 3'-O-methyltransferase (CalS11), an enzyme in calicheamicin biosynthesis.
Struct Dyn., 3, 2016
|
|
5JR3
 
 | | Crystal structure of carminomycin-4-O-methyltransferase DnrK in complex with SAH and 4-methylumbelliferone | | Descriptor: | 7-hydroxy-4-methyl-2H-chromen-2-one, Carminomycin 4-O-methyltransferase DnrK, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Wang, F, Johnson, B.R, Huber, T.D, Singh, S, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2016-05-05 | | Release date: | 2016-06-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of carminomycin-4-O-methyltransferase DnrK in complex with SAH and 4-methylumbelliferone (to be published)
To Be Published
|
|
5EEG
 
 | | Crystal structure of carminomycin-4-O-methyltransferase DnrK in complex with tetrazole-SAH | | Descriptor: | (2~{R},3~{R},4~{S},5~{S})-2-(6-aminopurin-9-yl)-5-[[(3~{S})-3-azanyl-3-(1~{H}-1,2,3,4-tetrazol-5-yl)propyl]sulfanylmethyl]oxolane-3,4-diol, Carminomycin 4-O-methyltransferase DnrK | | Authors: | Wang, F, Singh, S, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-10-22 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | Functional AdoMet Isosteres Resistant to Classical AdoMet Degradation Pathways.
Acs Chem.Biol., 11, 2016
|
|
1M7Q
 
 | | Crystal structure of p38 MAP kinase in complex with a dihydroquinazolinone inhibitor | | Descriptor: | 1-(2,6-DICHLOROPHENYL)-5-(2,4-DIFLUOROPHENYL)-7-PIPERAZIN-1-YL-3,4-DIHYDROQUINAZOLIN-2(1H)-ONE, Mitogen-activated protein kinase 14, SULFATE ION | | Authors: | Stelmach, J.E, Liu, L, Patel, S.B, Pivnichny, J.V, Scapin, G, Singh, S, Hop, C.E.C.A, Wang, Z, Cameron, P.M, Nichols, E.A, O'Keefe, S.J, O'Neill, E.A, Schmatz, D.M, Schwartz, C.D, Thompson, C.M, Zaller, D.M, Doherty, J.B. | | Deposit date: | 2002-07-22 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and synthesis of potent, orally bioavailable dihydroquinazolinone inhibitors of p38 MAP kinase.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
5EEH
 
 | | Crystal structure of carminomycin-4-O-methyltransferase DnrK in complex with SAH and 2-chloro-4-nitrophenol | | Descriptor: | 2-chloranyl-4-nitro-phenol, Carminomycin 4-O-methyltransferase DnrK, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Wang, F, Singh, S, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-10-22 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Functional AdoMet Isosteres Resistant to Classical AdoMet Degradation Pathways.
Acs Chem.Biol., 11, 2016
|
|
