4N7R
 
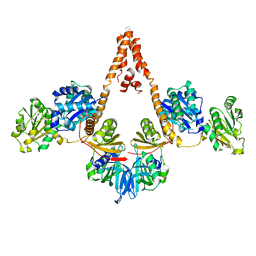 | | Crystal structure of Arabidopsis glutamyl-tRNA reductase in complex with its binding protein | | Descriptor: | Genomic DNA, chromosome 3, P1 clone: MXL8, ... | | Authors: | Zhao, A, Fang, Y, Lin, Y, Gong, W, Liu, L. | | Deposit date: | 2013-10-16 | | Release date: | 2014-05-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Crystal structure of Arabidopsis glutamyl-tRNA reductase in complex with its stimulator protein
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2N2T
 
 | | Solution NMR Structure of DE NOVO DESIGNED PROTEIN (FDA_60), Northeast Structural Genomics Consortium (NESG) Target OR303 | | Descriptor: | OR303 | | Authors: | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Hamilton, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-05-14 | | Release date: | 2015-09-16 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of DE NOVO DESIGNED PROTEIN (FDA_60), Northeast Structural Genomics Consortium (NESG) Target OR303
To be Published
|
|
2N3Z
 
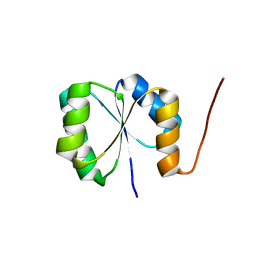 | | Solution NMR Structure of de novo designed protein, Rossmann2x2 Fold, Northeast Structural Genomics Consortium (NESG) Target OR446 | | Descriptor: | OR446 | | Authors: | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Pederson, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-15 | | Release date: | 2015-09-16 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of DE NOVO DESIGNED PROTEIN, Rossmann2x2 Fold, Northeast Structural Genomics Consortium (NESG) Target OR446
To be Published
|
|
2N2U
 
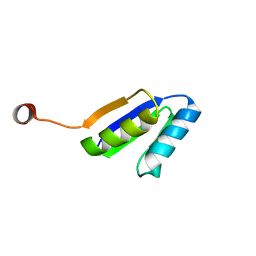 | | Solution NMR Structure of DE NOVO DESIGNED Ferredoxin Fold PROTEIN sfr3, Northeast Structural Genomics Consortium (NESG) Target OR358 | | Descriptor: | OR358 | | Authors: | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Hamilton, K, Pederson, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-05-14 | | Release date: | 2015-09-16 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of DE NOVO DESIGNED Ferredoxin Fold PROTEIN sfr3, Northeast Structural Genomics Consortium (NESG) Target OR358
To be Published
|
|
2N76
 
 | | Solution NMR Structure of De novo designed protein LFR1 1 with ferredoxin fold, Northeast Structural Genomics Consortium (NESG) Target OR414 | | Descriptor: | De novo designed protein LFR1 | | Authors: | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Pederson, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-09-03 | | Release date: | 2016-01-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of De novo designed protein LFR1 1 with ferredoxin fold, Northeast Structural Genomics Consortium (NESG) Target OR414
To be Published
|
|
2GHW
 
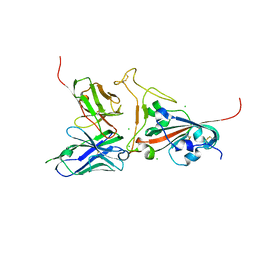 | | Crystal structure of SARS spike protein receptor binding domain in complex with a neutralizing antibody, 80R | | Descriptor: | CHLORIDE ION, Spike glycoprotein, anti-sars scFv antibody, ... | | Authors: | Hwang, W.C, Lin, Y, Santelli, E, Sui, J, Jaroszewski, L, Stec, B, Farzan, M, Marasco, W.A, Liddington, R.C. | | Deposit date: | 2006-03-27 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of neutralization by a human anti-severe acute respiratory syndrome spike protein antibody, 80R.
J.Biol.Chem., 281, 2006
|
|
2GHV
 
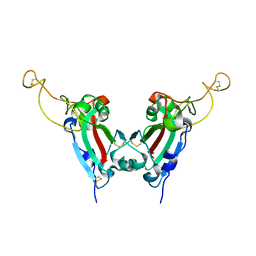 | | Crystal structure of SARS spike protein receptor binding domain | | Descriptor: | Spike glycoprotein | | Authors: | Hwang, W.C, Lin, Y, Santelli, E, Sui, J, Jaroszewski, L, Stec, B, Farzan, M, Marasco, W.A, Liddington, R.C. | | Deposit date: | 2006-03-27 | | Release date: | 2006-09-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of neutralization by a human anti-severe acute respiratory syndrome spike protein antibody, 80R.
J.Biol.Chem., 281, 2006
|
|
2N75
 
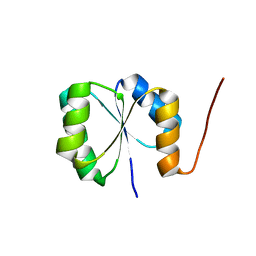 | | Solution NMR Structure of De novo designed protein, Rossmann2x2 Fold, Northeast Structural Genomics Consortium (NESG) Target OR446 | | Descriptor: | De novo designed protein | | Authors: | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Pederson, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-09-03 | | Release date: | 2016-01-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of De novo designed protein, Rossmann2x2 Fold, Northeast Structural Genomics Consortium (NESG) Target OR446
To be Published
|
|
3SHB
 
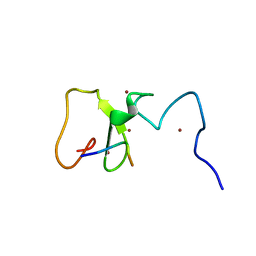 | | Crystal Structure of PHD Domain of UHRF1 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Histone H3 peptide, ZINC ION | | Authors: | Hu, L, Li, Z, Wang, P, Lin, Y, Xu, Y. | | Deposit date: | 2011-06-16 | | Release date: | 2011-08-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of PHD domain of UHRF1 and insights into recognition of unmodified histone H3 arginine residue 2.
Cell Res., 2011
|
|
8IMZ
 
 | | Cryo-EM structure of mouse Piezo1-MDFIC complex (composite map) | | Descriptor: | MyoD family inhibitor domain-containing protein, Piezo-type mechanosensitive ion channel component 1 | | Authors: | Zhou, Z, Ma, X, Lin, Y, Cheng, D, Bavi, N, Li, J.V, Sutton, D, Yao, M, Harvey, N, Corry, B, Zhang, Y, Cox, C.D. | | Deposit date: | 2023-03-07 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | MyoD-family inhibitor proteins act as auxiliary subunits of Piezo channels.
Science, 381, 2023
|
|
5W3N
 
 | | Molecular structure of FUS low sequence complexity domain protein fibrils | | Descriptor: | RNA-binding protein FUS | | Authors: | Murray, D.T, Kato, M, Lin, Y, Thurber, K, Hung, I, McKnight, S, Tycko, R. | | Deposit date: | 2017-06-08 | | Release date: | 2017-09-27 | | Last modified: | 2023-06-14 | | Method: | SOLID-STATE NMR | | Cite: | Structure of FUS Protein Fibrils and Its Relevance to Self-Assembly and Phase Separation of Low-Complexity Domains.
Cell, 171, 2017
|
|
4MDR
 
 | | Crystal structure of adaptor protein complex 4 (AP-4) mu4 subunit C-terminal domain D190A mutant, in complex with a sorting peptide from the amyloid precursor protein (APP) | | Descriptor: | AP-4 complex subunit mu-1, Amyloid beta A4 protein | | Authors: | Ross, B.H, Lin, Y, Corales, E.A, Burgos, P.V, Mardones, G.A. | | Deposit date: | 2013-08-23 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Functional Characterization of Cargo-Binding Sites on the mu 4-Subunit of Adaptor Protein Complex 4.
Plos One, 9, 2014
|
|
4QAU
 
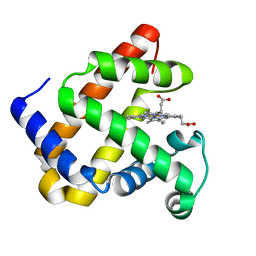 | | Crystal structure of F43Y mutant of sperm whale myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lin, Y, Tan, X, Li, W. | | Deposit date: | 2014-05-06 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | A Novel Tyrosine-Heme C-O Covalent Linkage in F43Y Myoglobin: A New Post-translational Modification of Heme Proteins
Chembiochem, 16, 2015
|
|
4LPI
 
 | |
7C96
 
 | | Avr1d:GmPUB13 U-box | | Descriptor: | RING-type E3 ubiquitin transferase, RxLR effector protein Avh6 | | Authors: | Xing, W, Hu, Q, Zhou, J, Yao, D. | | Deposit date: | 2020-06-05 | | Release date: | 2021-03-17 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Phytophthora sojae effector Avr1d functions as an E2 competitor and inhibits ubiquitination activity of GmPUB13 to facilitate infection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2FM4
 
 | |
3N9M
 
 | | ceKDM7A from C.elegans, alone | | Descriptor: | FE (II) ION, Putative uncharacterized protein, ZINC ION | | Authors: | Yang, Y, Hu, L, Wang, P, Hou, H, Chen, C.D, Xu, Y. | | Deposit date: | 2010-05-31 | | Release date: | 2010-06-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Structural insights into a dual-specificity histone demethylase ceKDM7A from Caenorhabditis elegans
Cell Res., 20, 2010
|
|
3N9P
 
 | | ceKDM7A from C.elegans, complex with H3K4me3K27me2 peptide and NOG | | Descriptor: | FE (II) ION, Histone H3 peptide, N-OXALYLGLYCINE, ... | | Authors: | Yang, Y, Hu, L, Wang, P, Hou, H, Chen, C.D, Xu, Y. | | Deposit date: | 2010-05-31 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.388 Å) | | Cite: | Structural insights into a dual-specificity histone demethylase ceKDM7A from Caenorhabditis elegans
Cell Res., 20, 2010
|
|
3N9O
 
 | | ceKDM7A from C.elegans, complex with H3K4me3 peptide, H3K9me2 peptide and NOG | | Descriptor: | FE (II) ION, Histone H3 peptide, N-OXALYLGLYCINE, ... | | Authors: | Yang, Y, Hu, L, Wang, P, Hou, H, Chen, C.D, Xu, Y. | | Deposit date: | 2010-05-31 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.309 Å) | | Cite: | Structural insights into a dual-specificity histone demethylase ceKDM7A from Caenorhabditis elegans
Cell Res., 20, 2010
|
|
3N9Q
 
 | | ceKDM7A from C.elegans, complex with H3K4me3 peptide, H3K27me2 peptide and NOG | | Descriptor: | FE (II) ION, Histone H3 peptide, N-OXALYLGLYCINE, ... | | Authors: | Yang, Y, Hu, L, Wang, P, Hou, H, Chen, C.D, Xu, Y. | | Deposit date: | 2010-05-31 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into a dual-specificity histone demethylase ceKDM7A from Caenorhabditis elegans
Cell Res., 20, 2010
|
|
3N9N
 
 | | ceKDM7A from C.elegans, complex with H3K4me3K9me2 peptide and NOG | | Descriptor: | FE (II) ION, Histone H3 peptide, N-OXALYLGLYCINE, ... | | Authors: | Yang, Y, Hu, L, Wang, P, Hou, H, Chen, C.D, Xu, Y. | | Deposit date: | 2010-05-31 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structural insights into a dual-specificity histone demethylase ceKDM7A from Caenorhabditis elegans
Cell Res., 20, 2010
|
|
3N9L
 
 | | ceKDM7A from C.elegans, complex with H3K4me3 peptide and NOG | | Descriptor: | FE (II) ION, Histone H3 peptide, N-OXALYLGLYCINE, ... | | Authors: | Yang, Y, Hu, L, Wang, P, Hou, H, Chen, C.D, Xu, Y. | | Deposit date: | 2010-05-31 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Structural insights into a dual-specificity histone demethylase ceKDM7A from Caenorhabditis elegans
Cell Res., 20, 2010
|
|
8WGV
 
 | | BA.2(S375) Spike (S6P)/hACE2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Wei, X, Zhang, Z. | | Deposit date: | 2023-09-22 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | The receptor binding domain of SARS-CoV-2 Omicron subvariants targets Siglec-9 to decrease its immunogenicity by preventing macrophage phagocytosis.
Nat.Immunol., 25, 2024
|
|
8WGW
 
 | | Local refinement of RBD-ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Wei, X, Zhang, Z. | | Deposit date: | 2023-09-22 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The receptor binding domain of SARS-CoV-2 Omicron subvariants targets Siglec-9 to decrease its immunogenicity by preventing macrophage phagocytosis.
Nat.Immunol., 25, 2024
|
|
6QNN
 
 | |
