3TBE
 
 | |
3RBJ
 
 | |
3RCC
 
 | |
3RBK
 
 | |
3RBI
 
 | |
3TB7
 
 | |
7LCG
 
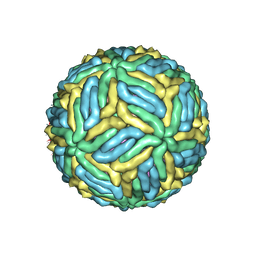 | | The mature Usutu SAAR-1776, Model A | | Descriptor: | (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, ... | | Authors: | Khare, B, Klose, T, Fang, Q, Kuhn, R. | | Deposit date: | 2021-01-11 | | Release date: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Structure of Usutu virus SAAR-1776 displays fusion loop asymmetry.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LCH
 
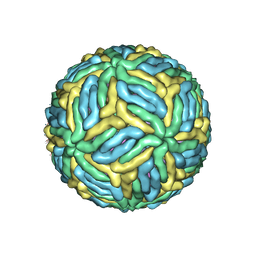 | | The mature Usutu SAAR-1776, Model B | | Descriptor: | (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, ... | | Authors: | Khare, B, Klose, T, Fang, Q, Kuhn, R. | | Deposit date: | 2021-01-11 | | Release date: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Structure of Usutu virus SAAR-1776 displays fusion loop asymmetry.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5V5I
 
 | |
5V5H
 
 | |
5V5G
 
 | |
1YHS
 
 | | Crystal structure of Pim-1 bound to staurosporine | | Descriptor: | Proto-oncogene serine/threonine-protein kinase Pim-1, STAUROSPORINE | | Authors: | Jacobs, M.D, Black, J, Futer, O, Swenson, L, Hare, B, Fleming, M, Saxena, K. | | Deposit date: | 2005-01-10 | | Release date: | 2005-01-25 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Pim-1 ligand-bound structures reveal the mechanism of serine/threonine kinase inhibition by LY294002.
J.Biol.Chem., 280, 2005
|
|
1YI3
 
 | | Crystal Structure of Pim-1 bound to LY294002 | | Descriptor: | 2-MORPHOLIN-4-YL-7-PHENYL-4H-CHROMEN-4-ONE, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Jacobs, M.D, Black, J, Futer, O, Swenson, L, Hare, B, Fleming, M, Saxena, K. | | Deposit date: | 2005-01-11 | | Release date: | 2005-01-25 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Pim-1 ligand-bound structures reveal the mechanism of serine/threonine kinase inhibition by LY294002.
J.Biol.Chem., 280, 2005
|
|
1YI4
 
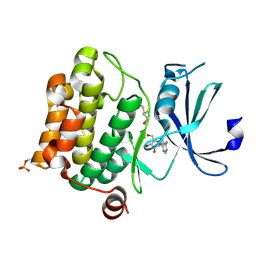 | | Structure of Pim-1 bound to adenosine | | Descriptor: | ADENOSINE, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Jacobs, M.D, Black, J, Futer, O, Swenson, L, Hare, B, Fleming, M, Saxena, K. | | Deposit date: | 2005-01-11 | | Release date: | 2005-01-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Pim-1 ligand-bound structures reveal the mechanism of serine/threonine kinase inhibition by LY294002.
J.Biol.Chem., 280, 2005
|
|
1HBW
 
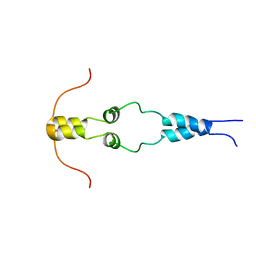 | | Solution nmr structure of the dimerization domain of the yeast transcriptional activator Gal4 (residues 50-106) | | Descriptor: | REGULATORY PROTEIN GAL4 | | Authors: | Hidalgo, P, Ansari, A.Z, Schmidt, P, Hare, B, Simkovic, N, Farrell, S, Shin, E.J, Ptashne, M, Wagner, G. | | Deposit date: | 2001-04-20 | | Release date: | 2001-05-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Recruitment of the Transcriptional Machinery Through Gal11P: Structure and Interactions of the GAL4 Dimerization Domain
Genes Dev., 15, 2001
|
|
1D9K
 
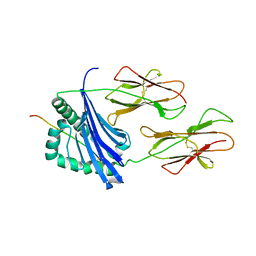 | | CRYSTAL STRUCTURE OF COMPLEX BETWEEN D10 TCR AND PMHC I-AK/CA | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CONALBUMIN PEPTIDE, ... | | Authors: | Reinherz, E.L, Tan, K, Tang, L, Kern, P, Liu, J.-H, Xiong, Y, Hussey, R.E, Smolyar, A, Hare, B, Zhang, R, Joachimiak, A, Chang, H.-C, Wagner, G, Wang, J.-H. | | Deposit date: | 1999-10-28 | | Release date: | 1999-12-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of a T cell receptor in complex with peptide and MHC class II.
Science, 286, 1999
|
|
8FMU
 
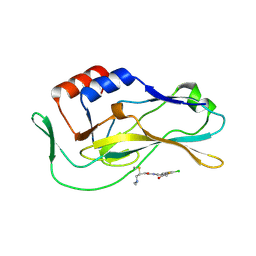 | | Crystal structure of human Brachyury G177D variant in complex with SJF-4601 | | Descriptor: | N-(3-chloro-4-fluorophenyl)-3-[4-(dimethylamino)butanamido]-4-methoxybenzamide, T-box transcription factor T | | Authors: | Bebenek, A, Linhares, B, Jaime-Figueroa, S, Butrin, A, Crews, C. | | Deposit date: | 2022-12-24 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery and Development of the First Selective Brachyury Degrader
To Be Published
|
|
6BXH
 
 | | Menin in complex with MI-853 | | Descriptor: | 1-{2-[4-(fluoroacetyl)piperazin-1-yl]ethyl}-4-methyl-5-[(4-{[6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidin-4-yl]amino}piperidin-1-yl)methyl]-1H-indole-2-carbonitrile, DIMETHYL SULFOXIDE, Menin, ... | | Authors: | Borkin, D, Klossowski, S, Pollock, J, Linhares, B, Cierpicki, T, Grembecka, J. | | Deposit date: | 2017-12-18 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.445 Å) | | Cite: | Menin in complex with MI-853
To Be Published
|
|
6C3N
 
 | | Crystal structure of BCL6 BTB domain in complex with compound 7CC5 | | Descriptor: | B-cell lymphoma 6 protein, N-(2-phenylethyl)-N'-pyridin-3-ylthiourea | | Authors: | Linhares, B, Cheng, H, Xue, F, Cierpicki, T. | | Deposit date: | 2018-01-10 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.53170586 Å) | | Cite: | Identification of Thiourea-Based Inhibitors of the B-Cell Lymphoma 6 BTB Domain via NMR-Based Fragment Screening and Computer-Aided Drug Design.
J.Med.Chem., 61, 2018
|
|
6C3L
 
 | | Crystal structure of BCL6 BTB domain with compound 15f | | Descriptor: | B-cell lymphoma 6 protein, N-[2-(1H-indol-3-yl)ethyl]-N'-{3-[(4-methylpiperazin-1-yl)methyl]-1-[2-(morpholin-4-yl)-2-oxoethyl]-1H-indol-6-yl}thiourea | | Authors: | Linhares, B, Cheng, H, Cierpicki, T, Xue, F. | | Deposit date: | 2018-01-10 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.46092153 Å) | | Cite: | Identification of Thiourea-Based Inhibitors of the B-Cell Lymphoma 6 BTB Domain via NMR-Based Fragment Screening and Computer-Aided Drug Design.
J.Med.Chem., 61, 2018
|
|
6BXY
 
 | | Menin in complex with MI-1481 | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-1-{[(2S)-5-oxomorpholin-2-yl]methyl}-5-[(4-{[6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidin-4-yl]amino}piperidin-1-yl)methyl]-1H-indole-2-carbonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Borkin, D, Klossowski, S, Pollock, J, Linhares, B, Cierpicki, T, Grembecka, J. | | Deposit date: | 2017-12-19 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Complexity of Blocking Bivalent Protein-Protein Interactions: Development of a Highly Potent Inhibitor of the Menin-Mixed-Lineage Leukemia Interaction.
J.Med.Chem., 61, 2018
|
|
6BY8
 
 | | Menin in complex with MI-1482 | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-1-{[(2R)-5-oxomorpholin-2-yl]methyl}-5-[(4-{[6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidin-4-yl]amino}piperidin-1-yl)methyl]-1H-indole-2-carbonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Borkin, T, Klossowski, S, Pollock, J, Linhares, B, Cierpicki, T, Grembecka, J. | | Deposit date: | 2017-12-20 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Complexity of Blocking Bivalent Protein-Protein Interactions: Development of a Highly Potent Inhibitor of the Menin-Mixed-Lineage Leukemia Interaction.
J.Med.Chem., 61, 2018
|
|
6POO
 
 | |
5V6A
 
 | |
5V69
 
 | |
