3QZZ
 
 | | 3D Structure of Ferric Methanosarcina Acetivorans Protoglobin Y61W mutant in Aquomet form | | Descriptor: | Methanosarcina acetivorans protoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pesce, A, Tilleman, L, Dewilde, S, Ascenzi, P, Coletta, M, Ciaccio, C, Bruno, S, Moens, L, Bolognesi, M, Nardini, M. | | Deposit date: | 2011-03-07 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural heterogeneity and ligand gating in ferric methanosarcina acetivorans protoglobin mutants.
Iubmb Life, 63, 2011
|
|
5CKA
 
 | | Human beta-2 microglobulin double mutant W60G-N83V | | Descriptor: | ACETATE ION, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sala, B.M, De Rosa, M, Bolognesi, M, Ricagno, S. | | Deposit date: | 2015-07-15 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational design of mutations that change the aggregation rate of a protein while maintaining its native structure and stability.
Sci Rep, 6, 2016
|
|
5CKG
 
 | | Human beta-2 microglobulin mutant V85E | | Descriptor: | ACETATE ION, Beta-2-microglobulin, GLYCEROL | | Authors: | Sala, B.M, De Rosa, M, Bolognesi, M, Ricagno, S. | | Deposit date: | 2015-07-15 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Rational design of mutations that change the aggregation rate of a protein while maintaining its native structure and stability.
Sci Rep, 6, 2016
|
|
3R0G
 
 | | 3D Structure of Ferric Methanosarcina Acetivorans Protoglobin I149F mutant in Aquomet form | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Pesce, A, Tilleman, L, Dewilde, S, Ascenzi, P, Coletta, M, Ciaccio, C, Bruno, S, Moens, L, Bolognesi, M, Nardini, M. | | Deposit date: | 2011-03-08 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural heterogeneity and ligand gating in ferric methanosarcina acetivorans protoglobin mutants.
Iubmb Life, 63, 2011
|
|
5CFH
 
 | |
3QZX
 
 | | 3D Structure of ferric methanosarcina acetivorans protoglobin Y61A mutant with unknown ligand | | Descriptor: | GLYCEROL, Methanosarcina acetivorans protoglobin, PHOSPHATE ION, ... | | Authors: | Pesce, A, Tilleman, L, Dewilde, S, Ascenzi, P, Coletta, M, Ciaccio, C, Bruno, S, Moens, L, Bolognesi, M, Nardini, M. | | Deposit date: | 2011-03-07 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural heterogeneity and ligand gating in ferric methanosarcina acetivorans protoglobin mutants.
Iubmb Life, 63, 2011
|
|
5CSG
 
 | |
5CSB
 
 | | The crystal structure of beta2-microglobulin D76N mutant at room temperature | | Descriptor: | Beta-2-microglobulin | | Authors: | de Rosa, M, Mota, C.S, de Sanctis, D, Bolognesi, M, Ricagno, S. | | Deposit date: | 2015-07-23 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.719 Å) | | Cite: | Conformational dynamics in crystals reveal the molecular bases for D76N beta-2 microglobulin aggregation propensity.
Nat Commun, 9, 2018
|
|
5CS7
 
 | | The crystal structure of wt beta2-microglobulin at room temperature | | Descriptor: | Beta-2-microglobulin | | Authors: | de Rosa, M, Mota, C.S, de Sanctis, D, Bolognesi, M, Ricagno, S. | | Deposit date: | 2015-07-23 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational dynamics in crystals reveal the molecular bases for D76N beta-2 microglobulin aggregation propensity.
Nat Commun, 9, 2018
|
|
1LHS
 
 | | LOGGERHEAD SEA TURTLE MYOGLOBIN (AQUO-MET) | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nardini, M, Tarricone, C, Lania, A, Desideri, A, De Sanctis, G, Coletta, M, Petruzzelli, R, Ascenzi, P, Coda, A, Bolognesi, M. | | Deposit date: | 1995-02-01 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reptile heme protein structure: X-ray crystallographic study of the aquo-met and cyano-met derivatives of the loggerhead sea turtle (Caretta caretta) myoglobin at 2.0 A resolution.
J.Mol.Biol., 247, 1995
|
|
1LHT
 
 | | LOGGERHEAD SEA TURTLE MYOGLOBIN (CYANO-MET) | | Descriptor: | CYANIDE ION, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nardini, M, Tarricone, C, Lania, A, Desideri, A, De Sanctis, G, Coletta, M, Petruzzelli, R, Ascenzi, P, Coda, A, Bolognesi, M. | | Deposit date: | 1995-02-01 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reptile heme protein structure: X-ray crystallographic study of the aquo-met and cyano-met derivatives of the loggerhead sea turtle (Caretta caretta) myoglobin at 2.0 A resolution.
J.Mol.Biol., 247, 1995
|
|
2BK9
 
 | | Drosophila Melanogaster globin | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, CG9734-PA, CHLORIDE ION, ... | | Authors: | de Sanctis, D, Dewilde, S, Pesce, A, Moens, L, Ascenzi, P, Hankeln, T, Burmester, T, Ponassi, M, Nardini, M, Bolognesi, M. | | Deposit date: | 2005-02-14 | | Release date: | 2005-05-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Bishistidyl Heme Hexacoordination, a Key Structural Property in Drosophila Melanogaster Hemoglobin
J.Biol.Chem., 280, 2005
|
|
2XYK
 
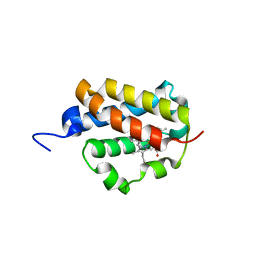 | | Group II 2-on-2 Hemoglobin from the Plant Pathogen Agrobacterium tumefaciens | | Descriptor: | 2-ON-2 HEMOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pesce, A, Nardini, M, LaBarre, M, Richard, C, Wittenberg, J.B, Wittenberg, B.A, Guertin, M, Bolognesi, M. | | Deposit date: | 2010-11-18 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Characterization of a Group II 2/2 Hemoglobin from the Plant Pathogen Agrobacterium Tumefaciens.
Biochim.Biophys.Acta, 1814, 2011
|
|
5GDS
 
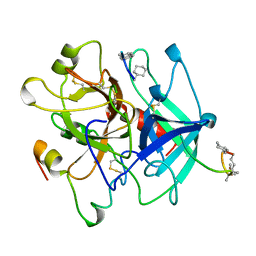 | | HIRUNORMS ARE TRUE HIRUDIN MIMETICS. THE CRYSTAL STRUCTURE OF HUMAN ALPHA-THROMBIN:HIRUNORM V COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-THROMBIN, HIRUNORM V | | Authors: | De Simone, G, Lombardi, A, Galdiero, S, Nastri, F, Della Morte, R, Staiano, N, Pedone, C, Bolognesi, M, Pavone, V. | | Deposit date: | 1997-07-17 | | Release date: | 1998-01-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Hirunorms are true hirudin mimetics. The crystal structure of human alpha-thrombin-hirunorm V complex.
Protein Sci., 7, 1998
|
|
1AVD
 
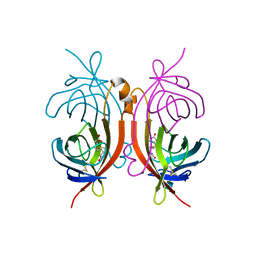 | | THREE-DIMENSIONAL STRUCTURE OF THE TETRAGONAL CRYSTAL FORM OF EGG-WHITE AVIDIN IN ITS FUNCTIONAL COMPLEX WITH BIOTIN AT 2.7 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AVIDIN, BIOTIN | | Authors: | Pugliese, L, Coda, A, Malcovati, M, Bolognesi, M. | | Deposit date: | 1993-03-05 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Three-dimensional structure of the tetragonal crystal form of egg-white avidin in its functional complex with biotin at 2.7 A resolution.
J.Mol.Biol., 231, 1993
|
|
1AVE
 
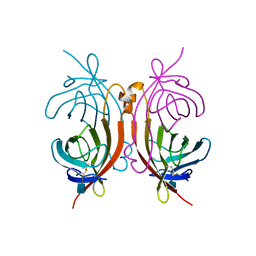 | | CRYSTAL STRUCTURE OF HEN EGG-WHITE APO-AVIDIN IN RELATION TO ITS THERMAL STABILITY PROPERTIES | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AVIDIN | | Authors: | Pugliese, L, Coda, A, Malcovati, M, Bolognesi, M. | | Deposit date: | 1993-03-05 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of apo-avidin from hen egg-white.
J.Mol.Biol., 235, 1994
|
|
1Y9O
 
 | | 1H NMR Structure of Acylphosphatase from the hyperthermophile Sulfolobus Solfataricus | | Descriptor: | Acylphosphatase | | Authors: | Corazza, A, Rosano, C, Pagano, K, Alverdi, V, Esposito, G, Capanni, C, Bemporad, F, Plakoutsi, G, Stefani, M, Chiti, F, Zuccotti, S, Bolognesi, M, Viglino, P. | | Deposit date: | 2004-12-16 | | Release date: | 2005-11-29 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure, conformational stability, and enzymatic properties of acylphosphatase from the hyperthermophile Sulfolobus solfataricus
Proteins, 62, 2006
|
|
5MUH
 
 | | Crystal structure of an amyloidogenic light chain dimer H7 | | Descriptor: | light chain dimer | | Authors: | Oberti, L, Rognoni, P, Russo, R, Maritan, M, Bacarizo, J, Bolognesi, M, Ricagno, S. | | Deposit date: | 2017-01-13 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
5MUD
 
 | | Crystal structure of an amyloidogenic light chain dimer H6 | | Descriptor: | light chain dimer,IGL@ protein | | Authors: | Oberti, L, Bacarizo, J, Maritan, M, Rognoni, P, Bolognesi, M, Ricagno, S. | | Deposit date: | 2017-01-13 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
4KL1
 
 | | HCN4 CNBD in complex with cGMP | | Descriptor: | ACETATE ION, CYCLIC GUANOSINE MONOPHOSPHATE, GLYCEROL, ... | | Authors: | Lolicato, M, Arrigoni, C, Zucca, S, Nardini, M, Bucchi, A, Schroeder, I, Simmons, K, Bolognesi, M, DiFrancesco, D, Schwede, F, Fishwick, C.W.G, Johnson, A.P.K, Thiel, G, Moroni, A. | | Deposit date: | 2013-05-07 | | Release date: | 2014-04-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cyclic dinucleotides bind the C-linker of HCN4 to control channel cAMP responsiveness.
Nat.Chem.Biol., 10, 2014
|
|
3DHJ
 
 | | Beta 2 microglobulin mutant W60C | | Descriptor: | Beta-2-microglobulin | | Authors: | Ricagno, S, Colombo, M, de Rosa, M, Bolognesi, M, Giorgetti, S, Bellotti, V. | | Deposit date: | 2008-06-18 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DE loop mutations affect beta2-microglobulin stability and amyloid aggregation
Biochem.Biophys.Res.Commun., 377, 2008
|
|
1YIO
 
 | | Crystallographic structure of response regulator StyR from Pseudomonas fluorescens | | Descriptor: | MAGNESIUM ION, MERCURY (II) ION, response regulatory protein | | Authors: | Milani, M, Leoni, L, Rampioni, G, Zennaro, E, Ascenzi, P, Bolognesi, M. | | Deposit date: | 2005-01-12 | | Release date: | 2005-09-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An Active-like Structure in the Unphosphorylated StyR Response Regulator Suggests a Phosphorylation- Dependent Allosteric Activation Mechanism
STRUCTURE, 13, 2005
|
|
5M6A
 
 | | Crystal structure of cardiotoxic Bence-Jones light chain dimer H9 | | Descriptor: | Bence-Jones light chain, GLYCEROL, PHOSPHATE ION | | Authors: | Oberti, L, Rognoni, P, Bacarizo, J, Bolognesi, M, Ricagno, S. | | Deposit date: | 2016-10-24 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
5M76
 
 | | Crystal structure of cardiotoxic Bence-Jones light chain dimer H10 | | Descriptor: | BROMIDE ION, light chain dimer | | Authors: | Oberti, L, Rognoni, P, Bacarizo, J, Bolognesi, M, Ricagno, S. | | Deposit date: | 2016-10-26 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
5M6I
 
 | | Crystal structure of non-cardiotoxic Bence-Jones light chain dimer M8 | | Descriptor: | SODIUM ION, light chain dimer | | Authors: | Oberti, L, Rognoni, P, Russo, R, Bacarizo, J, Bolognesi, M, Ricagno, S. | | Deposit date: | 2016-10-25 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
