4L68
 
 | |
4ZH1
 
 | | Complement factor H in complex with the GM1 glycan | | Descriptor: | Complement C3, Complement factor H, GLYCEROL, ... | | Authors: | Blaum, B.S, Stehle, T. | | Deposit date: | 2015-04-24 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Complement Factor H and Simian Virus 40 bind the GM1 ganglioside in distinct conformations.
Glycobiology, 26, 2016
|
|
4ONT
 
 | | Ternary host recognition complex of complement factor H, C3d, and sialic acid | | Descriptor: | Complement C3d fragment, Complement factor H, GLYCEROL, ... | | Authors: | Blaum, B.S, Stehle, T.S. | | Deposit date: | 2014-01-29 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for sialic acid-mediated self-recognition by complement factor H.
Nat.Chem.Biol., 11, 2015
|
|
6ZLZ
 
 | |
6ZML
 
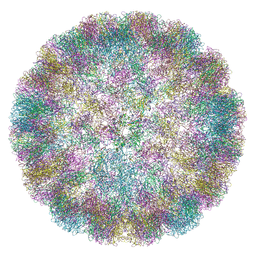 | | CryoEM Structure of Merkel Cell Polyomavirus Virus-like Particle | | Descriptor: | Capsid protein VP1 | | Authors: | Bayer, N.J, Januliene, D, Stehle, T, Moeller, A, Blaum, B.S. | | Deposit date: | 2020-07-03 | | Release date: | 2020-08-05 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of Merkel Cell Polyomavirus Capsid and Interaction with Its Glycosaminoglycan Attachment Receptor.
J.Virol., 94, 2020
|
|
3OXU
 
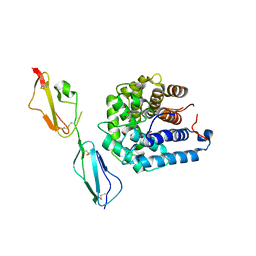 | | Complement components factor H CCP19-20 and C3d in complex | | Descriptor: | Complement C3, GLYCEROL, HF protein | | Authors: | Morgan, H.P, Schmidt, C.Q, Guariento, M, Gillespie, D, Herbert, A.P, Mertens, H, Blaum, B.S, Svergun, D, Johansson, C.M, Uhrin, D, Barlow, P.N, Hannan, J.P. | | Deposit date: | 2010-09-22 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for engagement by complement factor H of C3b on a self surface.
Nat.Struct.Mol.Biol., 18, 2011
|
|
6H9V
 
 | |
2JGX
 
 | | Structure of CCP module 7 of complement factor H - The AMD Not at risk varient (402Y) | | Descriptor: | COMPLEMENT FACTOR H | | Authors: | Herbert, A.P, Deakin, J.A, Schmidt, C.Q, Blaum, B.S, Egan, C, Ferreira, V.P, Pangburn, M.K, Lyon, M, Uhrin, D, Barlow, P.N. | | Deposit date: | 2007-02-16 | | Release date: | 2007-03-20 | | Last modified: | 2018-05-02 | | Method: | SOLUTION NMR | | Cite: | Structure shows that a glycosaminoglycan and protein recognition site in factor H is perturbed by age-related macular degeneration-linked single nucleotide polymorphism.
J. Biol. Chem., 282, 2007
|
|
2JGW
 
 | | Structure of CCP module 7 of complement factor H - The AMD at risk varient (402H) | | Descriptor: | COMPLEMENT FACTOR H | | Authors: | Herbert, A.P, Deakin, J.A, Schmidt, C.Q, Blaum, B.S, Egan, C, Ferreira, V.P, Pangburn, M.K, Lyon, M, Uhrin, D, Barlow, P.N. | | Deposit date: | 2007-02-16 | | Release date: | 2007-03-20 | | Last modified: | 2018-05-02 | | Method: | SOLUTION NMR | | Cite: | Structure shows that a glycosaminoglycan and protein recognition site in factor H is perturbed by age-related macular degeneration-linked single nucleotide polymorphism.
J. Biol. Chem., 282, 2007
|
|
6EQ9
 
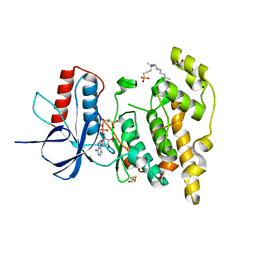 | | Crystal structure of JNK3 in complex with AMP-PCP | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Macedo, J.T, Stehle, T, Blaum, B.S. | | Deposit date: | 2017-10-12 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Optimization of a Pyridinylimidazole Scaffold: Shifting the Selectivity from p38 alpha Mitogen-Activated Protein Kinase to c-Jun N-Terminal Kinase 3.
ACS Omega, 3, 2018
|
|
6EKD
 
 | | Crystal structure of JNK3 in complex with a pyridinylimidazole inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-methyl-2-methylsulfanyl-1~{H}-imidazol-5-yl)-~{N}-(4-morpholin-4-ylphenyl)pyridin-2-amine, BETA-MERCAPTOETHANOL, ... | | Authors: | Macedo, J.T, Stehle, T, Blaum, B.S. | | Deposit date: | 2017-09-26 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Optimization of a Pyridinylimidazole Scaffold: Shifting the Selectivity from p38 alpha Mitogen-Activated Protein Kinase to c-Jun N-Terminal Kinase 3.
ACS Omega, 3, 2018
|
|
6EMH
 
 | | Crystal structure of JNK3 in complex with a pyridinylimidazole inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-methyl-1~{H}-imidazol-5-yl)-~{N}-(4-morpholin-4-ylphenyl)pyridin-2-amine, BETA-MERCAPTOETHANOL, ... | | Authors: | Macedo, J.T, Stehle, T, Blaum, B.S. | | Deposit date: | 2017-10-02 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Optimization of a Pyridinylimidazole Scaffold: Shifting the Selectivity from p38 alpha Mitogen-Activated Protein Kinase to c-Jun N-Terminal Kinase 3.
ACS Omega, 3, 2018
|
|
2UWN
 
 | | Crystal structure of Human Complement Factor H, SCR domains 6-8 (H402 risk variant), in complex with ligand. | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, ACETATE ION, CHLORIDE ION, ... | | Authors: | Prosser, B.E, Johnson, S, Roversi, P, Herbert, A.P, Blaum, B.S, Tyrrell, J, Jowitt, T.A, Clark, S.J, Terelli, E, Uhrin, D, Barlow, P.N, Sim, R.B, Day, A.J, Lea, S.M. | | Deposit date: | 2007-03-22 | | Release date: | 2007-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis for Complement Factor H Linked Age-Related Macular Degeneration.
J.Exp.Med., 204, 2007
|
|
2V8E
 
 | | Crystal structure of Human Complement Factor H, SCR domains 6-8 (H402 risk variant), in complex with ligand. | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, CHLORIDE ION, COMPLEMENT FACTOR H, ... | | Authors: | Prosser, B.E, Johnson, S, Roversi, P, Herbert, A.P, Blaum, B.S, Tyrrell, J, Jowitt, T.A, Clark, S.J, Tarelli, E, Uhrin, D, Barlow, P.N, Sim, R.B, Day, A.J, Lea, S.M. | | Deposit date: | 2007-08-07 | | Release date: | 2007-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Complement Factor H Linked Age-Related Macular Degeneration.
J.Exp.Med., 204, 2007
|
|
3RJ3
 
 | |
6G47
 
 | | Crystal Structure of Human Adenovirus 52 Short Fiber Knob in Complex with alpha-(2,8)-Trisialic Acid (DP3) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Liaci, A.M, Stehle, T. | | Deposit date: | 2018-03-26 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | Polysialic acid is a cellular receptor for human adenovirus 52.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6HKU
 
 | |
6HKV
 
 | |
4MJ0
 
 | | BK Polyomavirus VP1 pentamer in complex with GD3 oligosaccharide | | Descriptor: | CHLORIDE ION, GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid, ... | | Authors: | Neu, U, Stroh, L.J, Stehle, T. | | Deposit date: | 2013-09-03 | | Release date: | 2013-11-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Structure-Guided Mutation in the Major Capsid Protein Retargets BK Polyomavirus.
Plos Pathog., 9, 2013
|
|
4MJ1
 
 | | unliganded BK Polyomavirus VP1 pentamer | | Descriptor: | CHLORIDE ION, GLYCEROL, VP1 capsid protein | | Authors: | Neu, U, Stroh, L.J, Stehle, T. | | Deposit date: | 2013-09-03 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Structure-Guided Mutation in the Major Capsid Protein Retargets BK Polyomavirus.
Plos Pathog., 9, 2013
|
|
4FMG
 
 | |
4FMJ
 
 | | Merkel cell polyomavirus VP1 in complex with GD1a oligosaccharide | | Descriptor: | CHLORIDE ION, GLYCEROL, N-acetyl-alpha-neuraminic acid, ... | | Authors: | Neu, U, Hengel, H, Stehle, T. | | Deposit date: | 2012-06-17 | | Release date: | 2012-09-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of Merkel Cell Polyomavirus VP1 Complexes Define a Sialic Acid Binding Site Required for Infection.
Plos Pathog., 8, 2012
|
|
4FMH
 
 | | Merkel Cell Polyomavirus VP1 in complex with Disialyllactose | | Descriptor: | CHLORIDE ION, GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, ... | | Authors: | Neu, U, Hengel, H, Stehle, T. | | Deposit date: | 2012-06-17 | | Release date: | 2012-09-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of Merkel Cell Polyomavirus VP1 Complexes Define a Sialic Acid Binding Site Required for Infection.
Plos Pathog., 8, 2012
|
|
4FMI
 
 | | Merkel cell polyomavirus VP1 in complex with 3'-sialyllactosamine | | Descriptor: | CHLORIDE ION, GLYCEROL, N-acetyl-alpha-neuraminic acid, ... | | Authors: | Neu, U, Hengel, H, Stehle, T. | | Deposit date: | 2012-06-17 | | Release date: | 2012-09-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Merkel Cell Polyomavirus VP1 Complexes Define a Sialic Acid Binding Site Required for Infection.
Plos Pathog., 8, 2012
|
|
4GU3
 
 | |
