6IVU
 
 | |
6K4Y
 
 | | CryoEM structure of sigma appropriation complex | | Descriptor: | 10 kDa anti-sigma factor, DNA (60-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Shi, J, Wen, A, Feng, Y. | | Deposit date: | 2019-05-27 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structural basis of sigma appropriation.
Nucleic Acids Res., 47, 2019
|
|
5ULL
 
 | | CLOSTRIDIUM BEIJERINCKII FLAVODOXIN: REDUCED | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Ludwig, M.L, Pattridge, K.A, Metzger, A.L, Dixon, M.M, Eren, M, Feng, Y, Swenson, R. | | Deposit date: | 1997-01-09 | | Release date: | 1997-03-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Control of oxidation-reduction potentials in flavodoxin from Clostridium beijerinckii: the role of conformation changes.
Biochemistry, 36, 1997
|
|
6KJH
 
 | | Functional and structural insights into the unusual oxyanion hole-like geometry in macrolactin acyltransferase selective for dicarboxylic acyl donors | | Descriptor: | Putative beta-lactamase, SULFATE ION | | Authors: | Xiao, F, Sheng, D, Feng, Y, Li, W. | | Deposit date: | 2019-07-22 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural Basis of Specificity for Carboxyl-Terminated Acyl Donors in a Bacterial Acyltransferase.
J.Am.Chem.Soc., 142, 2020
|
|
6KJQ
 
 | | Functional and structural insights into the unusual oxyanion hole-like geometry in macrolactin acyltransferase selective for dicarboxylic acyl donors | | Descriptor: | (3~{Z},5~{E},8~{S},9~{E},11~{E},14~{S},16~{R},17~{Z},19~{E},24~{R})-24-methyl-8,14,16-tris(oxidanyl)-1-oxacyclotetracosa-3,5,9,11,17,19-hexaen-2-one, Putative beta-lactamase | | Authors: | Xiao, F, Dong, S, Feng, Y, Li, W. | | Deposit date: | 2019-07-23 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of Specificity for Carboxyl-Terminated Acyl Donors in a Bacterial Acyltransferase.
J.Am.Chem.Soc., 142, 2020
|
|
5VFK
 
 | |
6KFP
 
 | | Crystal structure of MavC ternary complex | | Descriptor: | MavC, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-conjugating enzyme E2 N | | Authors: | Mu, Y, Wang, Y, Han, Y, Li, D, Feng, Y. | | Deposit date: | 2019-07-08 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structural insights into the mechanism and inhibition of transglutaminase-induced ubiquitination by the Legionella effector MavC.
Nat Commun, 11, 2020
|
|
6KG6
 
 | | Crystal structure of MavC/UBE2N-Ub complex | | Descriptor: | MavC, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-conjugating enzyme E2 N | | Authors: | Wang, Y, Huang, Y, Chang, M, Feng, Y. | | Deposit date: | 2019-07-10 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural insights into the mechanism and inhibition of transglutaminase-induced ubiquitination by the Legionella effector MavC.
Nat Commun, 11, 2020
|
|
5F81
 
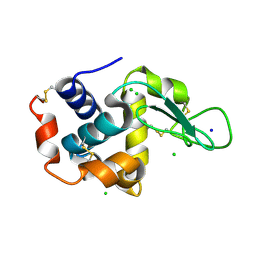 | | Acoustic injectors for drop-on-demand serial femtosecond crystallography | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Roessler, C.G, Agarwal, R, Allaire, M, Alonso-Mori, R, Andi, B, Bachega, J.F.R, Bommer, M, Brewster, A.S, Browne, M.C, Chatterjee, R, Cho, E, Cohen, A.E, Cowan, M, Datwani, S, Davidson, V.L, Defever, J, Eaton, B, Ellson, R, Feng, Y, Ghislain, L.P, Glownia, J.M, Han, G, Hattne, J, Hellmich, J, Heroux, A, Ibrahim, M, Kern, J, Kuczewski, A, Lemke, H.T, Liu, P, Majlof, L, McClintock, W.M, Myers, S, Nelsen, S, Olechno, J, Orville, A.M, Sauter, N.K, Soares, A.S, Soltis, M.S, Song, H, Stearns, R.G, Tran, R, Tsai, Y, Uervirojnangkoorn, M, Wilmot, C.M, Yachandra, V, Yano, J, Yukl, E.T, Zhu, D, Zouni, A. | | Deposit date: | 2015-12-08 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Acoustic Injectors for Drop-On-Demand Serial Femtosecond Crystallography.
Structure, 24, 2016
|
|
5HL4
 
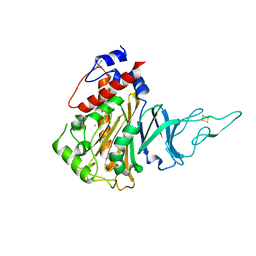 | | Acoustic injectors for drop-on-demand serial femtosecond crystallography | | Descriptor: | COBALT HEXAMMINE(III), FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Roessler, C.G, Agarwal, R, Allaire, M, Alonso-Mori, R, Andi, B, Bachega, J.F.R, Bommer, M, Brewster, A.S, Browne, M.C, Chatterjee, R, Cho, E, Cohen, A.E, Cowan, M, Datwani, S, Davidson, V.L, Defever, J, Eaton, B, Ellson, R, Feng, Y, Ghislain, L.P, Glownia, J.M, Han, G, Hattne, J, Hellmich, J, Heroux, A, Ibrahim, M, Kern, J, Kuczewski, A, Lemke, H.T, Liu, P, Majlof, L, McClintock, W.M, Myers, S, Nelsen, S, Olechno, J, Orville, A.M, Sauter, N.K, Soares, A.S, Soltis, M.S, Song, H, Stearns, R.G, Tran, R, Tsai, Y, Uervirojnangkoorn, M, Wilmot, C.M, Yachandra, V, Yano, J, Yukl, E.T, Zhu, D, Zouni, A. | | Deposit date: | 2016-01-14 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Acoustic Injectors for Drop-On-Demand Serial Femtosecond Crystallography.
Structure, 24, 2016
|
|
5HQD
 
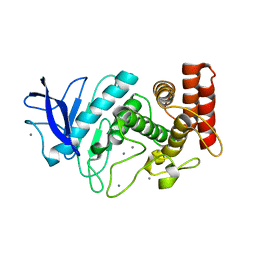 | | Acoustic injectors for drop-on-demand serial femtosecond crystallography | | Descriptor: | CALCIUM ION, Thermolysin, ZINC ION | | Authors: | Roesser, C.G, Agarwal, R, Allaire, M, Alonso-Mori, R, Andi, B, Bachega, J.F.R, Bommer, M, Brewster, A.S, Browne, M.C, Chatterjee, R, Cho, E, Cohen, A.E, Cowan, M, Datwani, S, Davidson, V.L, Defever, J, Eaton, B, Ellson, R, Feng, Y, Ghislain, L.P, Glownia, J.M, Han, G, Hattne, J, Hellmich, J, Heroux, A, Ibrahim, M, Kern, J, Kuczewski, A, Lemke, H.T, Liu, P, Majlof, L, McClintock, W.M, Myers, S, Nelsen, S, Olechno, J, Orville, A.M, Sauter, N.K, Soares, A.S, Soltis, M.S, Song, H, Stearns, R.G, Tran, R, Tsai, Y, Uervirojnangkoorn, M, Wilmot, C.M, Yachandra, V, Yano, J, Yukl, E.T, Zhu, D, Zouni, A. | | Deposit date: | 2016-01-21 | | Release date: | 2016-02-10 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Acoustic Injectors for Drop-On-Demand Serial Femtosecond Crystallography.
Structure, 24, 2016
|
|
5IAY
 
 | | NMR structure of UHRF1 Tandem Tudor Domains in a complex with Spacer peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Spacer | | Authors: | Fang, J, Cheng, J, Wang, J, Zhang, Q, Liu, M, Gong, R, Wang, P, Zhang, X, Feng, Y, Lan, W, Gong, Z, Tang, C, Wong, J, Yang, H, Cao, C, Xu, Y. | | Deposit date: | 2016-02-22 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Hemi-methylated DNA opens a closed conformation of UHRF1 to facilitate its histone recognition
Nat Commun, 7, 2016
|
|
3NCJ
 
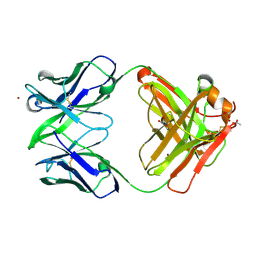 | | Crystal structure of Fab15 Mut8 | | Descriptor: | ACETATE ION, Fab15 Mut8 heavy chain, Fab15 Mut8 light chain, ... | | Authors: | Luo, J, Feng, Y, Gilliland, G.L. | | Deposit date: | 2010-06-04 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Co-evolution of antibody stability and Vk CDR-L3 canonical structure
To be Published
|
|
3NAA
 
 | | Crystal structure of Fab15 Mut5 | | Descriptor: | ACETATE ION, Fab15 Mut5 heavy chain, Fab15 Mut5 light chain, ... | | Authors: | Luo, J, Feng, Y, Gilliland, G.L. | | Deposit date: | 2010-06-01 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Co-evolution of antibody stability and Vk CDR-L3 canonical structure
To be Published
|
|
3NLL
 
 | | CLOSTRIDIUM BEIJERINCKII FLAVODOXIN MUTANT: G57A OXIDIZED | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Ludwig, M.L, Pattridge, K.A, Metzger, A.L, Dixon, M.M, Eren, M, Feng, Y, Swenson, R. | | Deposit date: | 1996-12-10 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Control of oxidation-reduction potentials in flavodoxin from Clostridium beijerinckii: the role of conformation changes.
Biochemistry, 36, 1997
|
|
5GQQ
 
 | | Structure of ALG-2/HEBP2 Complex | | Descriptor: | CALCIUM ION, CHLORIDE ION, Heme-binding protein 2, ... | | Authors: | Liu, X, Ma, J, Zhang, H, Feng, Y. | | Deposit date: | 2016-08-08 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Study of Apoptosis-linked Gene-2Heme-binding Protein 2 Interactions in HIV-1 Production.
J. Biol. Chem., 291, 2016
|
|
4MYD
 
 | | 1.37 Angstrom Crystal Structure of E. Coli 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase (MenH) in complex with SHCHC | | Descriptor: | 2-(3-CARBOXYPROPIONYL)-6-HYDROXY-CYCLOHEXA-2,4-DIENE CARBOXYLIC ACID, 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase | | Authors: | Sun, Y, Yin, S, Feng, Y, Li, J, Zhou, J, Liu, C, Zhu, G, Guo, Z. | | Deposit date: | 2013-09-27 | | Release date: | 2014-04-23 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.374 Å) | | Cite: | Molecular basis of the general base catalysis of an alpha / beta-hydrolase catalytic triad.
J.Biol.Chem., 289, 2014
|
|
4MXD
 
 | | 1.45 angstronm crystal structure of E.coli 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase (MenH) | | Descriptor: | 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Sun, Y, Yin, S, Feng, Y, Li, J, Zhou, J, Liu, C, Zhu, G, Guo, Z. | | Deposit date: | 2013-09-26 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular basis of the general base catalysis of an alpha / beta-hydrolase catalytic triad.
J.Biol.Chem., 289, 2014
|
|
6KYF
 
 | | Crystal structure of an anti-CRISPR protein | | Descriptor: | AcrF11, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Niu, Y, Wang, H, Zhang, Y, Feng, Y. | | Deposit date: | 2019-09-18 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | A Type I-F Anti-CRISPR Protein Inhibits the CRISPR-Cas Surveillance Complex by ADP-Ribosylation.
Mol.Cell, 80, 2020
|
|
2KQ3
 
 | | Solution structure of SNase140 | | Descriptor: | Thermonuclease | | Authors: | Wang, M, Feng, Y, Yao, H, Wang, J. | | Deposit date: | 2009-10-26 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Importance of the C-Terminal Loop L137-S141 for the Folding and Folding Stability of Staphylococcal Nuclease
Biochemistry, 49, 2010
|
|
2LCC
 
 | | Solution structure of RBBP1 chromobarrel domain | | Descriptor: | AT-rich interactive domain-containing protein 4A | | Authors: | Gong, W, Feng, Y. | | Deposit date: | 2011-04-28 | | Release date: | 2012-02-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insight into recognition of methylated histone tails by retinoblastoma-binding protein 1.
J.Biol.Chem., 2012
|
|
2M00
 
 | |
2MAM
 
 | |
2MBY
 
 | | NMR Structure of Rrp7 C-terminal Domain | | Descriptor: | Ribosomal RNA-processing protein 7 | | Authors: | Lin, J, Feng, Y, Ye, K. | | Deposit date: | 2013-08-08 | | Release date: | 2013-09-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An RNA-Binding Complex Involved in Ribosome Biogenesis Contains a Protein with Homology to tRNA CCA-Adding Enzyme.
Plos Biol., 11, 2013
|
|
2N51
 
 | |
