5TDH
 
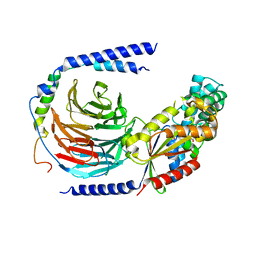 | | The crystal structure of the dominant negative mutant G protein alpha(i)-1-beta-1-gamma-2 G203A/A326S | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, P, Jia, M.-Z, Zhou, X.E, de Waal, P.W, Dickson, B.M, Liu, B, Hou, L, Yin, Y.-T, Kang, Y.-Y, Shi, Y, Melcher, K, Xu, H.E, Jiang, Y. | | Deposit date: | 2016-09-19 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structural basis of the dominant negative phenotype of the G alpha i1 beta 1 gamma 2 G203A/A326S heterotrimer
Acta Pharmacol.Sin., 37, 2016
|
|
7EY4
 
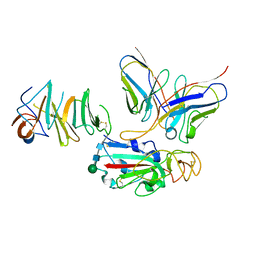 | | Local CryoEM of the SARS-CoV-2 S6PV2 in complex with BD-667 | | Descriptor: | BD-667 H, BD-667 L, Spike glycoprotein, ... | | Authors: | Liu, P.L. | | Deposit date: | 2021-05-29 | | Release date: | 2021-09-08 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Structures of SARS-CoV-2 B.1.351 neutralizing antibodies provide insights into cocktail design against concerning variants.
Cell Res., 31, 2021
|
|
7EZV
 
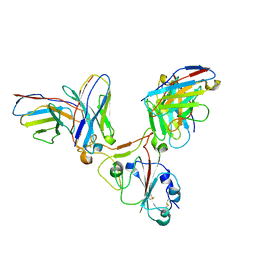 | |
2RCV
 
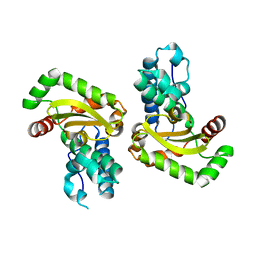 | | Crystal structure of the Bacillus subtilis superoxide dismutase | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase [Mn] | | Authors: | Liu, P, Ewis, H.E, Huang, Y.J, Lu, C.D, Tai, P.C, Weber, I.T. | | Deposit date: | 2007-09-20 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Bacillus subtilis superoxide dismutase.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
1TQH
 
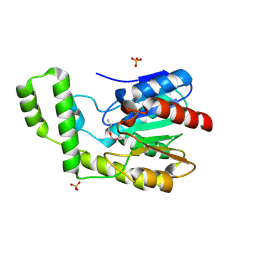 | | Covalent Reaction intermediate Revealed in Crystal Structure of the Geobacillus stearothermophilus Carboxylesterase Est30 | | Descriptor: | Carboxylesterase precursor, PROPYL ACETATE, SULFATE ION | | Authors: | Liu, P, Wang, Y.F, Ewis, H.E, Abdelal, A.T, Lu, C.D, Harrison, R.W, Weber, I.T. | | Deposit date: | 2004-06-17 | | Release date: | 2004-09-28 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Covalent reaction intermediate revealed in crystal structure of the Geobacillus stearothermophilus carboxylesterase Est30.
J.Mol.Biol., 342, 2004
|
|
2OGT
 
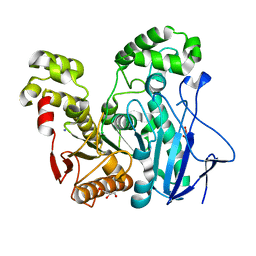 | | Crystal Structure of the Geobacillus Stearothermophilus Carboxylesterase EST55 at pH 6.8 | | Descriptor: | GLYCEROL, IODIDE ION, Thermostable carboxylesterase Est50 | | Authors: | Liu, P, Ewis, H.E, Tai, P.C, Lu, C.D, Weber, I.T. | | Deposit date: | 2007-01-08 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of the Geobacillus stearothermophilus Carboxylesterase Est55 and Its Activation of Prodrug CPT-11.
J.Mol.Biol., 367, 2007
|
|
2OGS
 
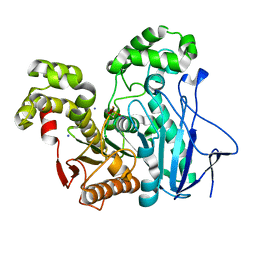 | | Crystal Structure of the GEOBACILLUS STEAROTHERMOPHILUS Carboxylesterase EST55 at pH 6.2 | | Descriptor: | IODIDE ION, Thermostable carboxylesterase Est50 | | Authors: | Liu, P, Ewis, H.E, Tai, P.C, Lu, C.D, Weber, I.T. | | Deposit date: | 2007-01-08 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal Structure of the Geobacillus stearothermophilus Carboxylesterase Est55 and Its Activation of Prodrug CPT-11.
J.Mol.Biol., 367, 2007
|
|
3U7U
 
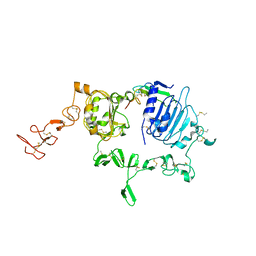 | | Crystal structure of extracellular region of human epidermal growth factor receptor 4 in complex with neuregulin-1 beta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neuregulin 1, Receptor tyrosine-protein kinase erbB-4 | | Authors: | Liu, P, Cleveland IV, T.E, Bouyain, S, Longo, P.A, Leahy, D.J. | | Deposit date: | 2011-10-14 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | A single ligand is sufficient to activate EGFR dimers.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7WRO
 
 | | Local structure of BD55-3372 and delta spike | | Descriptor: | 3372H, 3372L, Spike protein S1 | | Authors: | Liu, P.L. | | Deposit date: | 2022-01-27 | | Release date: | 2022-06-22 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
2R9B
 
 | | Structural Analysis of Plasmepsin 2 from Plasmodium falciparum complexed with a peptide-based inhibitor | | Descriptor: | Plasmepsin-2, peptide-based inhibitor | | Authors: | Liu, P, Marzahn, M.R, Robbins, A.H, McKenna, R, Dunn, B.M. | | Deposit date: | 2007-09-12 | | Release date: | 2007-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Recombinant plasmepsin 1 from the human malaria parasite plasmodium falciparum: enzymatic characterization, active site inhibitor design, and structural analysis.
Biochemistry, 48, 2009
|
|
3U2P
 
 | | Crystal structure of N-terminal three extracellular domains of ErbB4/Her4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor tyrosine-protein kinase erbB-4 | | Authors: | Liu, P, Bouyain, S, Elgenbrot, C, Leahy, D.J. | | Deposit date: | 2011-10-04 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The ErbB4 extracellular region retains a tethered-like conformation in the absence of the tether.
Protein Sci., 21, 2012
|
|
7WSC
 
 | | Local structure of BD55-3500 and omicron RBD complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3500H, 3500L, ... | | Authors: | Liu, P.L. | | Deposit date: | 2022-01-28 | | Release date: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Local structure of BD55-3500 and omicron RBD complex
To Be Published
|
|
7VHH
 
 | | Delta variant of SARS-CoV-2 Spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Liu, P. | | Deposit date: | 2021-09-22 | | Release date: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of delta variant SARS-CoV-2 Spike protein
To Be Published
|
|
1B0U
 
 | | ATP-BINDING SUBUNIT OF THE HISTIDINE PERMEASE FROM SALMONELLA TYPHIMURIUM | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, HISTIDINE PERMEASE | | Authors: | Hung, L.-W, Wang, I.X, Nikaido, K, Liu, P.-Q, Ames, G.F.-L, Kim, S.-H. | | Deposit date: | 1998-11-12 | | Release date: | 1999-11-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the ATP-binding subunit of an ABC transporter.
Nature, 396, 1998
|
|
1ZZB
 
 | | Crystal Structure of CoII HppE in Complex with Substrate | | Descriptor: | (S)-2-HYDROXYPROPYLPHOSPHONIC ACID, COBALT (II) ION, Hydroxypropylphosphonic Acid Epoxidase | | Authors: | Higgins, L.J, Yan, F, Liu, P, Liu, H.W, Drennan, C.L. | | Deposit date: | 2005-06-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insight into antibiotic fosfomycin biosynthesis by a mononuclear iron enzyme
Nature, 437, 2005
|
|
1ZZ8
 
 | | Crystal Structure of FeII HppE in Complex with Substrate Form 2 | | Descriptor: | (S)-2-HYDROXYPROPYLPHOSPHONIC ACID, FE (II) ION, Hydroxypropylphosphonic Acid Epoxidase | | Authors: | Higgins, L.J, Yan, F, Liu, P, Liu, H.W, Drennan, C.L. | | Deposit date: | 2005-06-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insight into antibiotic fosfomycin biosynthesis by a mononuclear iron enzyme
Nature, 437, 2005
|
|
1ZZ7
 
 | | Crystal Structure of FeII HppE in Complex with Substrate form 1 | | Descriptor: | (S)-2-HYDROXYPROPYLPHOSPHONIC ACID, FE (II) ION, Hydroxyprophylphosphonic Acid Epoxidase | | Authors: | Higgins, L.J, Yan, F, Liu, P, Liu, H.W, Drennan, C.L. | | Deposit date: | 2005-06-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insight into antibiotic fosfomycin biosynthesis by a mononuclear iron enzyme
Nature, 437, 2005
|
|
8W6P
 
 | | Crystal structure of dimeric murine SMPDL3A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acid sphingomyelinase-like phosphodiesterase 3a, ... | | Authors: | Zhang, C, Liu, P, Fan, S, Hou, Y. | | Deposit date: | 2023-08-29 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | SMPDL3A is a cGAMP-degrading enzyme induced by LXR-mediated lipid metabolism to restrict cGAS-STING DNA sensing.
Immunity, 56, 2023
|
|
1ZZC
 
 | | Crystal Structure of CoII HppE in Complex with Tris Buffer | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COBALT (II) ION, hydroxypropylphosphonic acid epoxidase | | Authors: | Higgins, L.J, Yan, F, Liu, P, Liu, H.W, Drennan, C.L. | | Deposit date: | 2005-06-13 | | Release date: | 2005-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into antibiotic fosfomycin biosynthesis by a mononuclear iron enzyme
Nature, 437, 2005
|
|
1ZZ6
 
 | | Crystal Structure of Apo-HppE | | Descriptor: | Hydroxypropylphosphonic Acid Epoxidase | | Authors: | Higgins, L.J, Yan, F, Liu, P, Liu, H.W, Drennan, C.L. | | Deposit date: | 2005-06-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into antibiotic fosfomycin biosynthesis by a mononuclear iron enzyme
Nature, 437, 2005
|
|
1ZZ9
 
 | | Crystal Structure of FeII HppE | | Descriptor: | FE (II) ION, Hydroxypropylphosphonic Acid Epoxidase, SULFATE ION | | Authors: | Higgins, L.J, Yan, F, Liu, P, Liu, H.W, Drennan, C.L. | | Deposit date: | 2005-06-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insight into antibiotic fosfomycin biosynthesis by a mononuclear iron enzyme
Nature, 437, 2005
|
|
1JXS
 
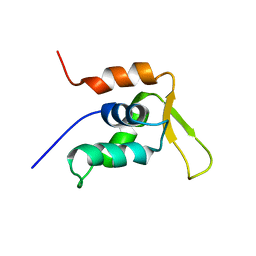 | | Solution Structure of the DNA-Binding Domain of Interleukin Enhancer Binding Factor | | Descriptor: | interleukin enhancer binding factor | | Authors: | Chuang, W.J, Liu, P.P, Li, C, Hsieh, Y.H, Chen, S.W, Chen, S.H, Jeng, W.Y. | | Deposit date: | 2001-09-08 | | Release date: | 2003-03-11 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA-binding domain of interleukin enhancer binding factor 1 (FOXK1a)
PROTEINS: STRUCT.,FUNCT.,GENET., 49, 2002
|
|
2VEW
 
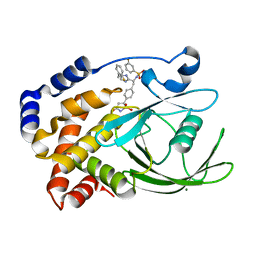 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B IN COMPLEX WITH AN ISOTHIAZOLIDINONE-CONTAINING INHIBITOR | | Descriptor: | 3-fluoro-N-[(1S)-1-[4-[(2-fluorophenyl)methyl]imidazol-2-yl]-2-[4-[(5S)-1,1,3-trioxo-1,2-thiazolidin-5-yl]phenyl]ethyl]benzenesulfonamide, MAGNESIUM ION, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 1 | | Authors: | Douty, B, Wayland, B, Ala, P.J, Bower, M.J, Pruitt, J, Bostrom, L, Wei, M, Klabe, R, Gonneville, L, Wynn, R, Burn, T.C, Liu, P.C.C, Combs, A.P, Yue, E.W. | | Deposit date: | 2007-10-27 | | Release date: | 2007-11-06 | | Last modified: | 2020-11-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Isothiazolidinone Inhibitors of Ptp1B Containing Imidazoles and Imidazolines
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2VEX
 
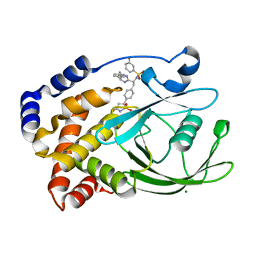 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B IN COMPLEX WITH AN ISOTHIAZOLIDINONE-CONTAINING INHIBITOR | | Descriptor: | MAGNESIUM ION, N-{(1S)-2-{4-[(5S)-1,1-dioxido-3-oxoisothiazolidin-5-yl]phenyl}-1-[(4R)-4-(2-phenylethyl)-4,5-dihydro-1H-imidazol-2-yl]ethyl}-3-fluorobenzenesulfonamide, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 1 | | Authors: | Douty, B, Wayland, B, Ala, P.J, Bower, M.J, Pruitt, J, Bostrom, L, Wei, M, Klabe, R, Gonneville, L, Wynn, R, Burn, T.C, Liu, P.C.C, Combs, A.P, Yue, E.W. | | Deposit date: | 2007-10-27 | | Release date: | 2007-11-06 | | Last modified: | 2020-11-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Isothiazolidinone Inhibitors of Ptp1B Containing Imidazoles and Imidazolines
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2VEV
 
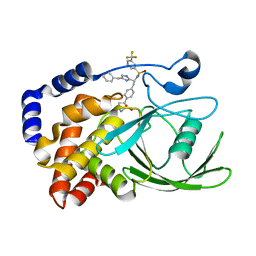 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B IN COMPLEX WITH AN ISOTHIAZOLIDINONE-CONTAINING INHIBITOR | | Descriptor: | MAGNESIUM ION, N-[(1S)-1-(4-benzyl-1H-imidazol-2-yl)-2-{4-[(5S)-1,1-dioxido-3-oxoisothiazolidin-5-yl]phenyl}ethyl]-3-(trifluoromethyl)benzenesulfonamide, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 1 | | Authors: | Douty, B, Wayland, B, Ala, P.J, Bower, M.J, Pruitt, J, Bostrom, L, Wei, M, Klabe, R, Gonneville, L, Wynn, R, Burn, T.C, Liu, P.C.C, Combs, A.P, Yue, E.W. | | Deposit date: | 2007-10-27 | | Release date: | 2007-11-06 | | Last modified: | 2020-11-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Isothiazolidinone Inhibitors of Ptp1B Containing Imidazoles and Imidazolines
Bioorg.Med.Chem.Lett., 18, 2008
|
|
