3OHN
 
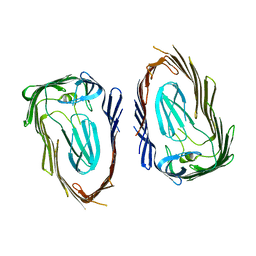 | | Crystal structure of the FimD translocation domain | | Descriptor: | Outer membrane usher protein FimD | | Authors: | Wang, T, Li, H. | | Deposit date: | 2010-08-17 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.011 Å) | | Cite: | Crystal structure of the FimD usher bound to its cognate FimC-FimH substrate.
Nature, 474, 2011
|
|
3SYJ
 
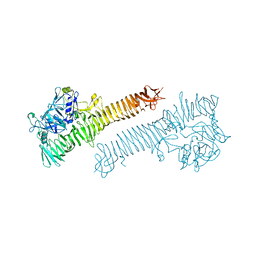 | | Crystal structure of the Haemophilus influenzae Hap adhesin | | Descriptor: | Adhesion and penetration protein autotransporter | | Authors: | Meng, G. | | Deposit date: | 2011-07-18 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the Haemophilus influenzae Hap adhesin reveals an intercellular oligomerization mechanism for bacterial aggregation
Embo J., 30, 2011
|
|
3ME0
 
 | | Structure of the E. coli chaperone PAPD in complex with the pilin domain of the PapGII adhesin | | Descriptor: | Chaperone protein papD, PapG protein | | Authors: | Ford, B.A, Elam, J.S, Dodson, K.W, Pinkner, J.S, Hultgren, S.J. | | Deposit date: | 2010-03-31 | | Release date: | 2011-04-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure of the E. Coli Chaperone PapD in Complex with the Pilin Domain of the PapGII Adhesin
To be Published
|
|
2MHJ
 
 | |
1HCT
 
 | |
1HCS
 
 | |
1KLF
 
 | | FIMH ADHESIN-FIMC CHAPERONE COMPLEX WITH D-MANNOSE | | Descriptor: | CHAPERONE PROTEIN FIMC, FIMH PROTEIN, alpha-D-mannopyranose | | Authors: | Hung, C.S, Bouckaert, J. | | Deposit date: | 2001-12-11 | | Release date: | 2002-06-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural basis of tropism of Escherichia coli to the bladder during urinary tract infection.
Mol.Microbiol., 44, 2002
|
|
1KIU
 
 | |
1AYD
 
 | |
1AYB
 
 | |
1AYA
 
 | |
1AYC
 
 | |
1FVJ
 
 | |
1FVK
 
 | |
3HK3
 
 | | Crystal structure of murine thrombin mutant W215A/E217A (one molecule in the asymmetric unit) | | Descriptor: | Thrombin heavy chain, Thrombin light chain | | Authors: | Gandhi, P.S, Page, M.J, Chen, Z, Bush-Pelc, L, Di Cera, E. | | Deposit date: | 2009-05-22 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanism of the Anticoagulant Activity of Thrombin Mutant W215A/E217A.
J.Biol.Chem., 284, 2009
|
|
3HK6
 
 | | Crystal structure of murine thrombin mutant W215A/E217A (two molecules in the asymmetric unit) | | Descriptor: | Thrombin heavy chain, Thrombin light chain | | Authors: | Gandhi, P.S, Page, M.J, Chen, Z, Bush-Pelc, L, Di Cera, E. | | Deposit date: | 2009-05-22 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Mechanism of the Anticoagulant Activity of Thrombin Mutant W215A/E217A.
J.Biol.Chem., 284, 2009
|
|
3HKI
 
 | | Crystal structure of murine thrombin mutant W215A/E217A in complex with the extracellular fragment of human PAR1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Proteinase-activated receptor 1, Thrombin heavy chain, ... | | Authors: | Gandhi, P.S, Page, M.J, Chen, Z, Bush-Pelc, L, Di Cera, E. | | Deposit date: | 2009-05-23 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of the Anticoagulant Activity of Thrombin Mutant W215A/E217A.
J.Biol.Chem., 284, 2009
|
|
3HKJ
 
 | | Crystal structure of human thrombin mutant W215A/E217A in complex with the extracellular fragment of human PAR1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Proteinase-activated receptor 1, Thrombin heavy chain, ... | | Authors: | Gandhi, P.S, Page, M.J, Chen, Z, Bush-Pelc, L, Di Cera, E. | | Deposit date: | 2009-05-23 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanism of the Anticoagulant Activity of Thrombin Mutant W215A/E217A.
J.Biol.Chem., 284, 2009
|
|
3AFP
 
 | | Crystal structure of the single-stranded DNA binding protein from Mycobacterium leprae (Form I) | | Descriptor: | CADMIUM ION, GLYCEROL, Single-stranded DNA-binding protein | | Authors: | Kaushal, P.S, Singh, P, Sharma, A, Muniyappa, K, Vijayan, M. | | Deposit date: | 2010-03-10 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray and molecular-dynamics studies on Mycobacterium leprae single-stranded DNA-binding protein and comparison with other eubacterial SSB structures
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3A5U
 
 | | Promiscuity and specificity in DNA binding to SSB: Insights from the structure of the Mycobacterium smegmatis SSB-ssDNA complex | | Descriptor: | DNA (31-MER), Single-stranded DNA-binding protein | | Authors: | Kaushal, P.S, Manjunath, G.P, Sekar, K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2009-08-12 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Promiscuity and specificity in DNA binding to SSB: Insights from the structure of the Mycobacterium smegmatis SSB-ssDNA complex.
To be Published, 2009
|
|
3AFQ
 
 | | Crystal structure of the single-stranded DNA binding protein from Mycobacterium leprae (Form II) | | Descriptor: | Single-stranded DNA-binding protein | | Authors: | Kaushal, P.S, Singh, P, Sharma, A, Muniyappa, K, Vijayan, M. | | Deposit date: | 2010-03-10 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray and molecular-dynamics studies on Mycobacterium leprae single-stranded DNA-binding protein and comparison with other eubacterial SSB structures
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1SKJ
 
 | | COCRYSTAL STRUCTURE OF UREA-SUBSTITUTED PHOSPHOPEPTIDE COMPLEX | | Descriptor: | 4-[3-CARBOXYMETHYL-3-(4-PHOSPHONOOXY-BENZYL)-UREIDO]-4-[(3-CYCLOHEXYL-PROPYL)-METHYL-CARBAMOYL]BUTYRIC ACID, PP60 V-SRC TYROSINE KINASE TRANSFORMING PROTEIN | | Authors: | Holland, D.R, Rubin, J.R. | | Deposit date: | 1997-09-18 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, synthesis, and cocrystal structure of a nonpeptide Src SH2 domain ligand.
J.Med.Chem., 40, 1997
|
|
1TCE
 
 | | SOLUTION NMR STRUCTURE OF THE SHC SH2 DOMAIN COMPLEXED WITH A TYROSINE-PHOSPHORYLATED PEPTIDE FROM THE T-CELL RECEPTOR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | PHOSPHOPEPTIDE OF THE ZETA CHAIN OF T CELL RECEPTOR, SHC | | Authors: | Zhou, M.-M, Meadows, R.P, Logan, T.M, Yoon, H.S, Wade, W.R, Ravichandran, K.S, Burakoff, S.J, Feisk, S.W. | | Deposit date: | 1996-03-27 | | Release date: | 1997-05-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Shc SH2 domain complexed with a tyrosine-phosphorylated peptide from the T-cell receptor.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1CL0
 
 | |
1BKL
 
 | | SELF-ASSOCIATED APO SRC SH2 DOMAIN | | Descriptor: | PP60 V-SRC TYROSINE KINASE TRANSFORMING PROTEIN | | Authors: | Holland, D.R, Rubin, J.R. | | Deposit date: | 1997-05-02 | | Release date: | 1997-07-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel Pp60Src Sh2 Domain Crystal Structures: A 2.0 Angstrom Co-Crystal Structure of a D-Amino Acid Substituted Phosphopeptide Complex and a 2.1 Angstrom Apo Structure Displaying Self-Association
To be Published
|
|
