5ULW
 
 | |
5WMB
 
 | | Structure of the 10S (-)-cis-BP-dG modified Rev1 ternary complex (the BP residue is disordered) | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Rechkoblit, O, Kolbanovsky, A, Landes, H, Geacintov, N.E, Aggarwal, A.K. | | Deposit date: | 2017-07-28 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanism of error-free replication across benzo[a]pyrene stereoisomers by Rev1 DNA polymerase.
Nat Commun, 8, 2017
|
|
5WM1
 
 | | Structure of the 10S (+)-trans-BP-dG modified Rev1 ternary complex | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, 1,2-ETHANEDIOL, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Rechkoblit, O, Kolbanovsky, A, Landes, H, Geacintov, N.E, Aggarwal, A.K. | | Deposit date: | 2017-07-28 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of error-free replication across benzo[a]pyrene stereoisomers by Rev1 DNA polymerase.
Nat Commun, 8, 2017
|
|
5WM8
 
 | | Structure of the 10R (+)-cis-BP-dG modified Rev1 ternary complex | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, 1,2-ETHANEDIOL, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Rechkoblit, O, Kolbanovsky, A, Landes, H, Geacintov, N.E, Aggarwal, A.K. | | Deposit date: | 2017-07-28 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Mechanism of error-free replication across benzo[a]pyrene stereoisomers by Rev1 DNA polymerase.
Nat Commun, 8, 2017
|
|
3PZP
 
 | | Human DNA polymerase kappa extending opposite a cis-syn thymine dimer | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*CP*CP*A)-3', 5'-D(*TP*TP*CP*CP*(TTD)P*GP*GP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Vasquez-Del Carpio, R, Silverstein, T.D, Lone, S, Johnson, R.E, Prakash, S, Prakash, L, Aggarwal, A.K. | | Deposit date: | 2010-12-14 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.336 Å) | | Cite: | Role of human DNA polymerase kappa in extension opposite from a cis-syn thymine dimer.
J.Mol.Biol., 408, 2011
|
|
3QU3
 
 | | Crystal structure of IRF-7 DBD apo form | | Descriptor: | 1,2-ETHANEDIOL, Interferon regulatory factor 7, SODIUM ION | | Authors: | De Ioannes, P.E, Escalante, C.R, Aggarwal, A.K. | | Deposit date: | 2011-02-23 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of apo IRF-3 and IRF-7 DNA binding domains: effect of loop L1 on DNA binding.
Nucleic Acids Res., 39, 2011
|
|
3QU6
 
 | | Crystal structure of IRF-3 DBD free form | | Descriptor: | CHLORIDE ION, IRF3 protein, SODIUM ION, ... | | Authors: | De Ioannes, P.E, Escalante, C.R, Aggarwal, A.K. | | Deposit date: | 2011-02-23 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of apo IRF-3 and IRF-7 DNA binding domains: effect of loop L1 on DNA binding.
Nucleic Acids Res., 39, 2011
|
|
3TQ1
 
 | | Human DNA Polymerase eta in binary complex with DNA | | Descriptor: | DNA (5'-D(*TP*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*TP*CP*AP*TP*TP*AP*TP*GP*AP*CP*GP*CP*T)-3'), DNA polymerase eta | | Authors: | Ummat, A, Silverstein, T.D, Jain, R, Buku, A, Johnson, R.E, Prakash, L, Prakash, S, Aggarwal, A.K. | | Deposit date: | 2011-09-08 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.556 Å) | | Cite: | Human DNA Polymerase Eta Is Pre-Aligned for dNTP Binding and Catalysis.
J.Mol.Biol., 415, 2012
|
|
3OHB
 
 | | Yeast DNA polymerase eta extending from an 8-oxoG lesion | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*GP*TP*CP*CP*TP*CP*CP*CP*CP*TP*(DOC))-3', 5'-D(*TP*AP*AP*TP*GP*(8OG)P*AP*GP*GP*GP*GP*AP*GP*GP*AP*C)-3', ... | | Authors: | Silverstein, T.D, Jain, R, Aggarwal, A.K. | | Deposit date: | 2010-08-17 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for error-free replication of oxidatively damaged DNA by yeast DNA polymerase eta.
Structure, 18, 2010
|
|
3MFI
 
 | | DNA Polymerase Eta in Complex With a cis-syn Thymidine Dimer | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(*GP*TP*CP*CP*TP*CP*CP*CP*CP*TP*(DOC))-3', 5'-D(*TP*AP*AP*(TTD)P*GP*AP*GP*GP*GP*GP*AP*GP*GP*AP*C)-3', ... | | Authors: | Silverstein, T.D, Johnson, R.E, Jain, R, Prakash, L, Prakash, S, Aggarwal, A.K. | | Deposit date: | 2010-04-02 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural basis for the suppression of skin cancers by DNA polymerase eta.
Nature, 465, 2010
|
|
3MFH
 
 | | DNA Polymerase Eta in Complex With Undamaged DNA | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(*GP*TP*CP*CP*TP*CP*CP*CP*CP*TP*(DOC))-3', 5'-D(*TP*AP*AP*TP*TP*GP*AP*GP*GP*GP*GP*AP*GP*GP*AP*C)-3', ... | | Authors: | Silverstein, T.D, Johnson, R.E, Jain, R, Prakash, L, Prakash, S, Aggarwal, A.K. | | Deposit date: | 2010-04-02 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the suppression of skin cancers by DNA polymerase eta.
Nature, 465, 2010
|
|
3OHA
 
 | | Yeast DNA polymerase eta inserting dCTP opposite an 8oxoG lesion | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*GP*TP*CP*CP*TP*CP*CP*CP*CP*TP*(DOC))-3', 5'-D(P*TP*(8OG)P*GP*AP*GP*GP*GP*GP*AP*GP*GP*AP*C)-3', ... | | Authors: | Silverstein, T.D, Jain, R, Aggarwal, A.K. | | Deposit date: | 2010-08-17 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for error-free replication of oxidatively damaged DNA by yeast DNA polymerase eta.
Structure, 18, 2010
|
|
3OSP
 
 | | Structure of rev1 | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*AP*TP*CP*CP*TP*CP*CP*CP*CP*TP*AP*(DOC))-3', 5'-D(*TP*AP*AP*(3DR)P*GP*TP*AP*GP*GP*GP*GP*AP*GP*GP*AP*T)-3', ... | | Authors: | Nair, D.T, Aggarwal, A.K. | | Deposit date: | 2010-09-09 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | DNA synthesis across an abasic lesion by yeast REV1 DNA polymerase.
J.Mol.Biol., 406, 2011
|
|
3ODH
 
 | | Structure of OkrAI/DNA complex | | Descriptor: | CALCIUM ION, DNA (5'-D(*TP*AP*TP*GP*GP*AP*TP*CP*CP*AP*TP*A)-3'), OkrAI endonuclease | | Authors: | Vanamee, E.S, Aggarwal, A.K. | | Deposit date: | 2010-08-11 | | Release date: | 2010-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Asymmetric DNA recognition by the OkrAI endonuclease, an isoschizomer of BamHI.
Nucleic Acids Res., 39, 2011
|
|
1FOK
 
 | | STRUCTURE OF RESTRICTION ENDONUCLEASE FOKI BOUND TO DNA | | Descriptor: | DNA (5'-D(*AP*TP*GP*AP*CP*TP*AP*GP*CP*GP*TP*TP*AP*TP*CP*AP*T P*CP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*GP*AP*TP*GP*AP*TP*AP*AP*CP*GP*CP*TP*AP*G P*TP*CP*A)-3'), PROTEIN (FOKI RESTRICTION ENDONUCLEASE) | | Authors: | Aggarwal, D.A, Wah, J.A, Hirsch, L.F, Dorner, I, Schildkraut, A.K. | | Deposit date: | 1997-04-18 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the multimodular endonuclease FokI bound to DNA.
Nature, 388, 1997
|
|
1IF1
 
 | | INTERFERON REGULATORY FACTOR 1 (IRF-1) COMPLEX WITH DNA | | Descriptor: | DNA (26-MER), PROTEIN (INTERFERON REGULATORY FACTOR 1) | | Authors: | Escalante, C.R, Yie, J, Thanos, D, Aggarwal, A. | | Deposit date: | 1997-09-12 | | Release date: | 1998-02-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of IRF-1 with bound DNA reveals determinants of interferon regulation.
Nature, 391, 1998
|
|
2LWD
 
 | |
2LWE
 
 | |
1ZET
 
 | |
1PER
 
 | |
1JM4
 
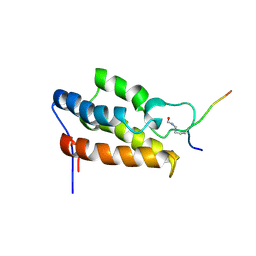 | | NMR Structure of P/CAF Bromodomain in Complex with HIV-1 Tat Peptide | | Descriptor: | HIV-1 Tat Peptide, P300/CBP-associated Factor | | Authors: | Mujtaba, S, He, Y, Zeng, L, Farooq, A, Carlson, J.E, Ott, M, Verdin, E, Zhou, M.-M. | | Deposit date: | 2001-07-17 | | Release date: | 2002-07-17 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of lysine-acetylated HIV-1 Tat recognition by PCAF bromodomain
Mol.Cell, 9, 2002
|
|
1LMB
 
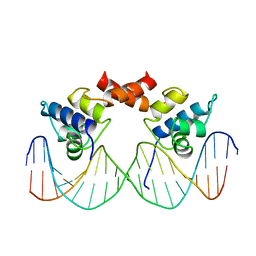 | | REFINED 1.8 ANGSTROM CRYSTAL STRUCTURE OF THE LAMBDA REPRESSOR-OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*CP*CP*AP*CP*TP*GP*GP*CP*GP*GP*TP*GP*A P*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*TP*CP*AP*CP*CP*GP*CP*CP*AP*GP*TP*GP*G P*TP*AP*T)-3'), PROTEIN (LAMBDA REPRESSOR) | | Authors: | Beamer, L.J, Pabo, C.O. | | Deposit date: | 1991-11-05 | | Release date: | 1991-11-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined 1.8 A crystal structure of the lambda repressor-operator complex.
J.Mol.Biol., 227, 1992
|
|
3CRO
 
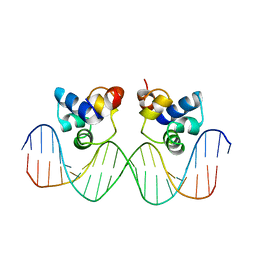 | | THE PHAGE 434 CRO/OR1 COMPLEX AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*AP*AP*GP*TP*AP*CP*AP*AP*AP*CP*TP*TP*TP*CP*TP*TP*G P*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*CP*AP*AP*GP*AP*AP*AP*GP*TP*TP*TP*GP*T P*AP*CP*T)-3'), PROTEIN (434 CRO) | | Authors: | Mondragon, A, Harrison, S.C. | | Deposit date: | 1990-07-06 | | Release date: | 1991-10-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The phage 434 Cro/OR1 complex at 2.5 A resolution.
J.Mol.Biol., 219, 1991
|
|
