3FHT
 
 | |
3FHC
 
 | | Crystal structure of human Dbp5 in complex with Nup214 | | Descriptor: | ATP-dependent RNA helicase DDX19B, Nuclear pore complex protein Nup214 | | Authors: | von Moeller, H, Conti, E. | | Deposit date: | 2008-12-09 | | Release date: | 2009-02-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner
Nat.Struct.Mol.Biol., 16, 2009
|
|
8RXB
 
 | |
4Q8G
 
 | |
4W8X
 
 | | Crystal Structure of Cmr1 from Pyrococcus furiosus bound to a nucleotide | | Descriptor: | CRISPR system Cmr subunit Cmr1-1, GUANOSINE-3'-MONOPHOSPHATE, PHOSPHATE ION | | Authors: | Benda, C, Ebert, J, Baumgaertner, M, Conti, E. | | Deposit date: | 2014-08-26 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Model of a CRISPR RNA-Silencing Complex Reveals the RNA-Target Cleavage Activity in Cmr4.
Mol.Cell, 56, 2014
|
|
4W8Y
 
 | | Structure of full length Cmr2 from Pyrococcus furiosus (Manganese bound form) | | Descriptor: | CRISPR system Cmr subunit Cmr2, MANGANESE (II) ION, ZINC ION | | Authors: | Benda, C, Ebert, J, Baumgaertner, M, Conti, E. | | Deposit date: | 2014-08-26 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Model of a CRISPR RNA-Silencing Complex Reveals the RNA-Target Cleavage Activity in Cmr4.
Mol.Cell, 56, 2014
|
|
4JHJ
 
 | | Crystal structure of Danio rerio Slip1 in complex with Dbp5 | | Descriptor: | DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 19 (DBP5 homolog, yeast), MIF4G domain-containing protein B | | Authors: | Von Moeller, H, Conti, E. | | Deposit date: | 2013-03-05 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural and biochemical studies of SLIP1-SLBP identify DBP5 and eIF3g as SLIP1-binding proteins.
Nucleic Acids Res., 41, 2013
|
|
7QDS
 
 | | Apo human SKI complex in the open state | | Descriptor: | Helicase SKI2W, Tetratricopeptide repeat protein 37, WD repeat-containing protein 61 | | Authors: | Koegel, A, Keidel, A, Bonneau, A, Schaefer, I.B, Conti, E. | | Deposit date: | 2021-11-30 | | Release date: | 2022-02-02 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The human SKI complex regulates channeling of ribosome-bound RNA to the exosome via an intrinsic gatekeeping mechanism.
Mol.Cell, 82, 2022
|
|
7QDR
 
 | | Apo human SKI complex in the closed state | | Descriptor: | Helicase SKI2W, Tetratricopeptide repeat protein 37, WD repeat-containing protein 61 | | Authors: | Koegel, A, Keidel, A, Bonneau, F, Schaefer, I.B, Conti, E. | | Deposit date: | 2021-11-29 | | Release date: | 2022-02-02 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The human SKI complex regulates channeling of ribosome-bound RNA to the exosome via an intrinsic gatekeeping mechanism.
Mol.Cell, 82, 2022
|
|
7QE0
 
 | | 80S-bound human SKI complex in the open state | | Descriptor: | Helicase SKI2W, RNA (5'-R(P*UP*UP*UP*UP*UP*UP*UP*UP*U)-3') | | Authors: | Koegel, A, Keidel, A, Bonneau, F, Schaefer, I.B, Conti, E. | | Deposit date: | 2021-12-01 | | Release date: | 2022-02-09 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | The human SKI complex regulates channeling of ribosome-bound RNA to the exosome via an intrinsic gatekeeping mechanism.
Mol.Cell, 82, 2022
|
|
7QDZ
 
 | | 80S-bound human SKI complex in the closed state | | Descriptor: | Helicase SKI2W, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3'), Tetratricopeptide repeat protein 37, ... | | Authors: | Koegel, A, Keidel, A, Bonneau, F, Schaefer, I.B, Conti, E. | | Deposit date: | 2021-12-01 | | Release date: | 2022-02-09 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The human SKI complex regulates channeling of ribosome-bound RNA to the exosome via an intrinsic gatekeeping mechanism.
Mol.Cell, 82, 2022
|
|
7QDY
 
 | | RNA-bound human SKI complex | | Descriptor: | Helicase SKI2W, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3'), Tetratricopeptide repeat protein 37, ... | | Authors: | Koegel, A, Keidel, A, Bonneau, F, Schaefer, I.B, Conti, E. | | Deposit date: | 2021-12-01 | | Release date: | 2022-02-09 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The human SKI complex regulates channeling of ribosome-bound RNA to the exosome via an intrinsic gatekeeping mechanism.
Mol.Cell, 82, 2022
|
|
4WFD
 
 | | Structure of the Rrp6-Rrp47-Mtr4 interaction | | Descriptor: | ATP-dependent RNA helicase DOB1, Exosome complex exonuclease RRP6, Exosome complex protein LRP1, ... | | Authors: | Schuch, B, Conti, E. | | Deposit date: | 2014-09-14 | | Release date: | 2014-10-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The exosome-binding factors Rrp6 and Rrp47 form a composite surface for recruiting the Mtr4 helicase.
Embo J., 33, 2014
|
|
4TXD
 
 | | Crystal structure of Thermofilum pendens Csc2 | | Descriptor: | Csc2, ZINC ION | | Authors: | Hrle, A, Conti, E. | | Deposit date: | 2014-07-03 | | Release date: | 2014-08-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.818 Å) | | Cite: | Structural analyses of the CRISPR protein Csc2 reveal the RNA-binding interface of the type I-D Cas7 family.
Rna Biol., 11, 2014
|
|
4WFC
 
 | | Structure of the Rrp6-Rrp47 interaction | | Descriptor: | Exosome complex exonuclease RRP6, Exosome complex protein LRP1, SULFATE ION | | Authors: | Schuch, B, Conti, E. | | Deposit date: | 2014-09-14 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The exosome-binding factors Rrp6 and Rrp47 form a composite surface for recruiting the Mtr4 helicase.
Embo J., 33, 2014
|
|
4U4C
 
 | | The molecular architecture of the TRAMP complex reveals the organization and interplay of its two catalytic activities | | Descriptor: | 1,2-ETHANEDIOL, ATP-dependent RNA helicase DOB1, CHLORIDE ION, ... | | Authors: | Falk, S, Weir, J.R, Hentschel, J, Reichelt, P, Bonneau, F, Conti, E. | | Deposit date: | 2014-07-23 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Molecular Architecture of the TRAMP Complex Reveals the Organization and Interplay of Its Two Catalytic Activities.
Mol.Cell, 55, 2014
|
|
4W8V
 
 | |
4W8W
 
 | |
4JHK
 
 | |
4ZRL
 
 | | Structure of the non canonical Poly(A) polymerase complex GLD-2 - GLD-3 | | Descriptor: | CHLORIDE ION, Defective in germ line development protein 3, Poly(A) RNA polymerase gld-2 | | Authors: | Nakel, K, Bonneau, F, Eckmann, C.R, Conti, E. | | Deposit date: | 2015-05-12 | | Release date: | 2015-07-08 | | Last modified: | 2017-09-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural basis for the activation of the C. elegans noncanonical cytoplasmic poly(A)-polymerase GLD-2 by GLD-3.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2BR2
 
 | | RNase PH core of the archaeal exosome | | Descriptor: | CHLORIDE ION, EXOSOME COMPLEX EXONUCLEASE 1, EXOSOME COMPLEX EXONUCLEASE 2 | | Authors: | lorentzen, E, Fribourg, S, Conti, E. | | Deposit date: | 2005-04-30 | | Release date: | 2005-06-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Archaeal Exosome Core is a Hexameric Ring Structure with Three Catalytic Subunits.
Nat.Struct.Mol.Biol., 12, 2005
|
|
2C37
 
 | | RNASE PH CORE OF THE ARCHAEAL EXOSOME IN COMPLEX WITH U8 RNA | | Descriptor: | 5-O-phosphono-beta-D-ribofuranose, CHLORIDE ION, PROBABLE EXOSOME COMPLEX EXONUCLEASE 1, ... | | Authors: | Lorentzen, E, Conti, E. | | Deposit date: | 2005-10-04 | | Release date: | 2005-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of 3' End RNA Recognition and Exoribonucleolytic Cleavage by an Exosome Rnase Ph Core.
Mol.Cell, 20, 2005
|
|
2C38
 
 | | RNase PH core of the archaeal exosome in complex with A5 RNA | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, PROBABLE EXOSOME COMPLEX EXONUCLEASE 1, ... | | Authors: | Lorentzen, E, Conti, E. | | Deposit date: | 2005-10-04 | | Release date: | 2005-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of 3' End RNA Recognition and Exoribonucleolytic Cleavage by an Exosome Rnase Ph Core.
Mol.Cell, 20, 2005
|
|
2C39
 
 | | RNase PH core of the archaeal exosome in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PROBABLE EXOSOME COMPLEX EXONUCLEASE 1, PROBABLE EXOSOME COMPLEX EXONUCLEASE 2 | | Authors: | Lorentzen, E, Conti, E. | | Deposit date: | 2005-10-05 | | Release date: | 2005-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Basis of 3' End RNA Recognition and Exoribonucleolytic Cleavage by an Exosome Rnase Ph Core.
Mol.Cell, 20, 2005
|
|
7PW4
 
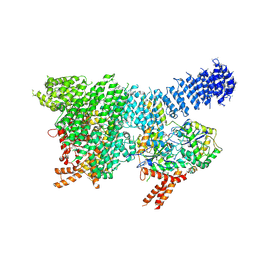 | | Human SMG1-8-9 kinase complex bound to a SMG1 inhibitor | | Descriptor: | 1-[4-[4-[2-[[4-chloranyl-3-(diethylsulfamoyl)phenyl]amino]pyrimidin-4-yl]pyridin-2-yl]phenyl]-3-methyl-urea, ADENOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Langer, L.M, Conti, E. | | Deposit date: | 2021-10-06 | | Release date: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Cryo-EM reconstructions of inhibitor-bound SMG1 kinase reveal an autoinhibitory state dependent on SMG8.
Elife, 10, 2021
|
|
