1YM1
 
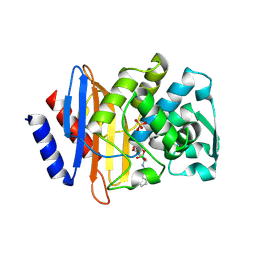 | | X-ray crystallographic structure of CTX-M-9 beta-lactamase complexed with a boronic acid inhibitor (SM2) | | Descriptor: | (1R)-1-(2-THIENYLACETYLAMINO)-1-(3-CARBOXYPHENYL)METHYLBORONIC ACID, PHOSPHATE ION, beta-lactamase CTX-M-9a | | Authors: | Chen, Y, Shoichet, B, Bonnet, R. | | Deposit date: | 2005-01-20 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structure, Function, and Inhibition along the Reaction Coordinate of CTX-M beta-Lactamases.
J.Am.Chem.Soc., 127, 2005
|
|
1YLW
 
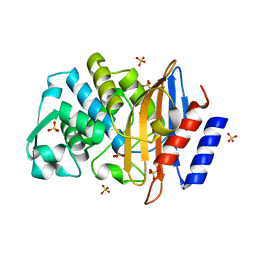 | | X-ray structure of CTX-M-16 beta-lactamase | | Descriptor: | CTX-M-16 beta-lactamase, SULFATE ION | | Authors: | Chen, Y, Delmas, J, Sirot, J, Shoichet, B, Bonnet, R. | | Deposit date: | 2005-01-19 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Atomic Resolution Structures of CTX-M beta-Lactamases: Extended Spectrum Activities from Increased Mobility and Decreased Stability.
J.Mol.Biol., 348, 2005
|
|
8UAQ
 
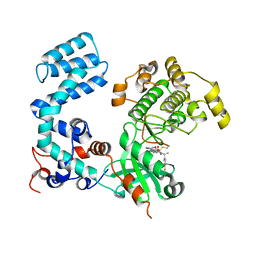 | | Crystal Structure of Human G Protein-Coupled Receptor Kinase 5 in Complex with GRL018-21 | | Descriptor: | (3Z)-N-[(1R)-1-(4-fluorophenyl)ethyl]-3-[(4-{[(2S)-2-(furan-2-yl)-2-hydroxyacetyl]amino}-3,5-dimethyl-1H-pyrrol-2-yl)methylidene]-2-oxo-2,3-dihydro-1H-indole-5-carboxamide, G protein-coupled receptor kinase 5 | | Authors: | Chen, Y, Tesmer, J.J.G. | | Deposit date: | 2023-09-21 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Development of a new class of potent and highly selective G protein-coupled receptor kinase 5 inhibitors and structural insight from crystal structures of inhibitor complexes.
Eur.J.Med.Chem., 264, 2023
|
|
8UAP
 
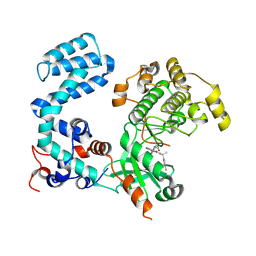 | | Crystal Structure of Human G Protein-Coupled Receptor Kinase 5 D311N in Complex with CCG273441 | | Descriptor: | (3Z)-3-{[4-(2-chloroacetamido)-3,5-dimethyl-1H-pyrrol-2-yl]methylidene}-N-[(1R)-1-(4-fluorophenyl)ethyl]-2-oxo-2,3-dihydro-1H-indole-5-carboxamide, G protein-coupled receptor kinase 5 | | Authors: | Chen, Y, Tesmer, J.J.G. | | Deposit date: | 2023-09-21 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Development of a new class of potent and highly selective G protein-coupled receptor kinase 5 inhibitors and structural insight from crystal structures of inhibitor complexes.
Eur.J.Med.Chem., 264, 2023
|
|
6DWS
 
 | | Crystal structure of human GRP78 in complex with (2R,3R,4S,5R)-2-(6-amino-8-((2-chlorobenzyl)amino)-9H-purin-9-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol | | Descriptor: | 8-{[(2-chlorophenyl)methyl]amino}adenosine, Endoplasmic reticulum chaperone BiP, MAGNESIUM ION | | Authors: | Chen, Y, Antoshchenko, T, Smil, D, Zepeda, C, Huang, Y, Park, H.W. | | Deposit date: | 2018-06-27 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human GRP78 in complex with (2R,3R,4S,5R)-2-(6-amino-8-((2-chlorobenzyl)amino)-9H-purin-9-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol
To Be Published
|
|
2XEX
 
 | | crystal structure of Staphylococcus aureus elongation factor G | | Descriptor: | CHLORIDE ION, ELONGATION FACTOR G, POTASSIUM ION | | Authors: | Chen, Y, Koripella, R.K, Sanyal, S, Selmer, M. | | Deposit date: | 2010-05-19 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Staphylococcus Aureus Elongation Factor G - Structure and Analysis of a Target for Fusidic Acid.
FEBS J., 277, 2010
|
|
4CS6
 
 | | Crystal structure of AadA - an aminoglycoside adenyltransferase | | Descriptor: | AMINOGLYCOSIDE ADENYLTRANSFERASE | | Authors: | Chen, Y, Nasvall, J, Andersson, D.I, Selmer, M. | | Deposit date: | 2014-03-05 | | Release date: | 2015-03-25 | | Last modified: | 2015-11-11 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structure of Aada from Salmonella Enterica: A Monomeric Aminoglycoside (3'')(9) Adenyltransferase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4IK5
 
 | | High resolution structure of Delta-REST-GCaMP3 | | Descriptor: | CALCIUM ION, RCaMP, Green fluorescent protein | | Authors: | Chen, Y, Song, X, Miao, L, Zhu, Y, Ji, G. | | Deposit date: | 2012-12-25 | | Release date: | 2014-01-29 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insight into enhanced calcium indicator GCaMP3 and GCaMPJ to promote further improvement.
Protein Cell, 4, 2013
|
|
4IK1
 
 | | High resolution structure of GCaMPJ at pH 8.5 | | Descriptor: | CALCIUM ION, RCaMP, Green fluorescent protein | | Authors: | Chen, Y, Song, X, Miao, L, Zhu, Y, Ji, G. | | Deposit date: | 2012-12-25 | | Release date: | 2014-02-05 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into enhanced calcium indicator GCaMP3 and GCaMPJ to promote further improvement.
Protein Cell, 4, 2013
|
|
4IK4
 
 | | High resolution structure of GCaMP3 at pH 5.0 | | Descriptor: | CALCIUM ION, RCaMP, Green fluorescent protein | | Authors: | Chen, Y, Song, X, Miao, L, Zhu, Y, Ji, G. | | Deposit date: | 2012-12-25 | | Release date: | 2014-02-05 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural insight into enhanced calcium indicator GCaMP3 and GCaMPJ to promote further improvement.
Protein Cell, 4, 2013
|
|
4CRU
 
 | | Complex of human CNOT9 and CNOT1 including one tryptophan | | Descriptor: | CCR4-NOT TRANSCRIPTION COMPLEX SUBUNIT 1, CELL DIFFERENTIATION PROTEIN RCD1 HOMOLOG, GLYCEROL | | Authors: | Chen, Y, Boland, A, Izaurralde, E, Weichenrieder, O. | | Deposit date: | 2014-03-01 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Ddx6-Cnot1 Complex and W-Binding Pockets in Cnot9 Reveal Direct Links between Mirna Target Recognition and Silencing
Mol.Cell, 54, 2014
|
|
4IK3
 
 | | High resolution structure of GCaMP3 at pH 8.5 | | Descriptor: | CALCIUM ION, RCaMP, Green fluorescent protein | | Authors: | Chen, Y, Song, X, Miao, L, Zhu, Y, Ji, G. | | Deposit date: | 2012-12-25 | | Release date: | 2014-01-29 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Structural insight into enhanced calcium indicator GCaMP3 and GCaMPJ to promote further improvement.
Protein Cell, 4, 2013
|
|
4CRW
 
 | | Complex of human DDX6 (RECA-C) and CNOT1 (MIF4G) | | Descriptor: | CCR4-NOT TRANSCRIPTION COMPLEX SUBUNIT 1, GLYCEROL, PROBABLE ATP-DEPENDENT RNA HELICASE DDX6 | | Authors: | Chen, Y, Boland, A, Izaurralde, E, Weichenrieder, O. | | Deposit date: | 2014-03-01 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Ddx6-Cnot1 Complex and W-Binding Pockets in Cnot9 Reveal Direct Links between Mirna Target Recognition and Silencing
Mol.Cell, 54, 2014
|
|
4IK9
 
 | | High resolution structure of GCaMP3 dimer form 2 at pH 7.5 | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, RCaMP, ... | | Authors: | Chen, Y, Song, X, Miao, L, Zhu, Y, Ji, G. | | Deposit date: | 2012-12-25 | | Release date: | 2014-01-29 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into enhanced calcium indicator GCaMP3 and GCaMPJ to promote further improvement.
Protein Cell, 4, 2013
|
|
4IK8
 
 | | High resolution structure of GCaMP3 dimer form 1 at pH 7.5 | | Descriptor: | CALCIUM ION, RCaMP, Green fluorescent protein | | Authors: | Chen, Y, Song, X, Miao, L, Zhu, Y, Ji, G. | | Deposit date: | 2012-12-25 | | Release date: | 2014-02-05 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insight into enhanced calcium indicator GCaMP3 and GCaMPJ to promote further improvement.
Protein Cell, 4, 2013
|
|
8I4B
 
 | | Cryo-EM structure of apo-form ABCC4 | | Descriptor: | ATP-binding cassette sub-family C member 4 | | Authors: | Chen, Y, Wang, L, Hou, W.T, Zhou, C.Z, Chen, Y, Li, Q. | | Deposit date: | 2023-01-19 | | Release date: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Cryo-EM structure ofABCC4
Nat Cardiovasc Res, 2023
|
|
8I4A
 
 | | Cryo-EM structure of dipyridamole-bound ABCC4 | | Descriptor: | 2-[[2-[bis(2-hydroxyethyl)amino]-4,8-di(piperidin-1-yl)pyrimido[5,4-d]pyrimidin-6-yl]-(2-hydroxyethyl)amino]ethanol, ATP-binding cassette sub-family C member 4 | | Authors: | Chen, Y, Wang, L, Hou, W.T, Zhou, C.Z, Chen, Y, Li, Q. | | Deposit date: | 2023-01-19 | | Release date: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure ofABCC4
Nat Cardiovasc Res, 2023
|
|
8I4C
 
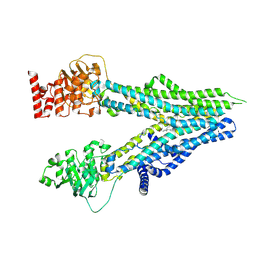 | | Cryo-EM structure of U46619-bound ABCC4 | | Descriptor: | (5Z)-7-{(1R,4S,5S,6R)-6-[(1E,3S)-3-hydroxyoct-1-en-1-yl]-2-oxabicyclo[2.2.1]hept-5-yl}hept-5-enoic acid, ATP-binding cassette sub-family C member 4 | | Authors: | Chen, Y, Wang, L, Hou, W.T, Zhou, C.Z, Chen, Y, Li, Q. | | Deposit date: | 2023-01-19 | | Release date: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Cryo-EM structure ofABCC4
Nat Cardiovasc Res, 2023
|
|
4L3N
 
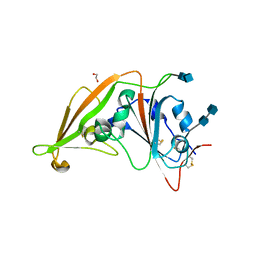 | | Crystal structure of the receptor-binding domain from newly emerged Middle East respiratory syndrome coronavirus | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | Authors: | Chen, Y, Rajashankar, K.R, Yang, Y, Agnihothram, S.S, Liu, C, Lin, Y.-L, Baric, R.S, Li, F. | | Deposit date: | 2013-06-06 | | Release date: | 2013-07-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of the receptor-binding domain from newly emerged middle East respiratory syndrome coronavirus.
J.Virol., 87, 2013
|
|
4FYP
 
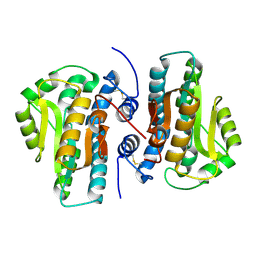 | | Crystal Structure of Plant Vegetative Storage Protein | | Descriptor: | MAGNESIUM ION, Vegetative storage protein 1 | | Authors: | Chen, Y, Wei, J, Wang, M, Gong, W, Zhang, M. | | Deposit date: | 2012-07-05 | | Release date: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of Arabidopsis VSP1 reveals the plant class C-like phosphatase structure of the DDDD superfamily of phosphohydrolases
Plos One, 7, 2012
|
|
3C7T
 
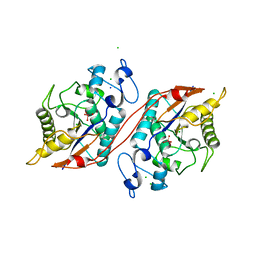 | | Crystal structure of the ecdysone phosphate phosphatase, EPPase, from Bombix mori in complex with tungstate | | Descriptor: | CHLORIDE ION, Ecdysteroid-phosphate phosphatase, IODIDE ION, ... | | Authors: | Chen, Y, Carpino, N, Nassar, N. | | Deposit date: | 2008-02-08 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and functional characterization of the c-terminal domain of the ecdysteroid phosphate phosphatase from Bombyx mori reveals a new enzymatic activity.
Biochemistry, 47, 2008
|
|
3D4I
 
 | |
3D6A
 
 | | Crystal structure of the 2H-phosphatase domain of Sts-2 in complex with tungstate. | | Descriptor: | MAGNESIUM ION, SODIUM ION, Sts-2 protein, ... | | Authors: | Chen, Y, Carpino, N, Nassar, N. | | Deposit date: | 2008-05-19 | | Release date: | 2009-03-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional characterization of the 2H-phosphatase domain of Sts-2 reveals an acid-dependent phosphatase activity.
Biochemistry, 48, 2009
|
|
5GTM
 
 | | Modified human MxA, nucleotide-free form | | Descriptor: | Interferon-induced GTP-binding protein Mx1 | | Authors: | Chen, Y, Gao, S. | | Deposit date: | 2016-08-22 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.896 Å) | | Cite: | Conformational dynamics of dynamin-like MxA revealed by single-molecule FRET
Nat Commun, 8, 2017
|
|
3FKW
 
 | | AmpC K67R mutant apo structure | | Descriptor: | Beta-lactamase, PHOSPHATE ION, POTASSIUM ION | | Authors: | Chen, Y, McReynolds, A, Shoichet, B.K. | | Deposit date: | 2008-12-17 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Re-examining the role of Lys67 in class C beta-lactamase catalysis.
Protein Sci., 18, 2009
|
|
