4X63
 
 | | Crystal structure of PRMT5:MEP50 with EPZ015666 and SAH | | Descriptor: | Methylosome protein 50, N-[(2S)-3-(3,4-dihydroisoquinolin-2(1H)-yl)-2-hydroxypropyl]-6-(oxetan-3-ylamino)pyrimidine-4-carboxamide, Protein arginine N-methyltransferase 5, ... | | Authors: | Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-06 | | Release date: | 2015-04-22 | | Last modified: | 2015-05-27 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models.
Nat.Chem.Biol., 11, 2015
|
|
4X60
 
 | | Crystal structure of PRMT5:MEP50 with EPZ015666 and sinefungin | | Descriptor: | GLYCEROL, Methylosome protein 50, N-[(2S)-3-(3,4-dihydroisoquinolin-2(1H)-yl)-2-hydroxypropyl]-6-(oxetan-3-ylamino)pyrimidine-4-carboxamide, ... | | Authors: | Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-06 | | Release date: | 2015-04-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models.
Nat.Chem.Biol., 11, 2015
|
|
4X61
 
 | | Crystal structure of PRMT5:MEP50 with EPZ015666 and SAM | | Descriptor: | GLYCEROL, Methylosome protein 50, N-[(2S)-3-(3,4-dihydroisoquinolin-2(1H)-yl)-2-hydroxypropyl]-6-(oxetan-3-ylamino)pyrimidine-4-carboxamide, ... | | Authors: | Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-06 | | Release date: | 2015-04-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models.
Nat.Chem.Biol., 11, 2015
|
|
6CEN
 
 | | Crystal Structure of WHSC1L1 in Complex with Inhibitor PEP21 | | Descriptor: | ACE-GLY-VAL-NLE-ARG-ILE-NH2, Histone-lysine N-methyltransferase NSD3, S-ADENOSYLMETHIONINE, ... | | Authors: | Boriack-Sjodin, P.A, Swinger, K, Farrow, N.A. | | Deposit date: | 2018-02-12 | | Release date: | 2018-05-09 | | Last modified: | 2018-05-23 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Identification of a peptide inhibitor for the histone methyltransferase WHSC1.
PLoS ONE, 13, 2018
|
|
3FAA
 
 | |
3EOT
 
 | |
3U2D
 
 | | S. aureus GyrB ATPase domain in complex with small molecule inhibitor | | Descriptor: | 4-bromo-5-methyl-N-[1-(3-nitropyridin-2-yl)piperidin-4-yl]-1H-pyrrole-2-carboxamide, DNA gyrase subunit B, MAGNESIUM ION | | Authors: | Boriack-Sjodin, P.A, Prince, D.B, Eakin, A.E, Sherer, B.A. | | Deposit date: | 2011-10-03 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Pyrrolamide DNA gyrase inhibitors: fragment-based nuclear magnetic resonance screening to identify antibacterial agents.
Antimicrob.Agents Chemother., 56, 2012
|
|
3U2K
 
 | | S. aureus GyrB ATPase domain in complex with a small molecule inhibitor | | Descriptor: | 2-chloro-6-(4-{[(3,4-dichloro-5-methyl-1H-pyrrol-2-yl)carbonyl]amino}piperidin-1-yl)pyridine-4-carboxamide, DNA gyrase subunit B, MAGNESIUM ION | | Authors: | Boriack-Sjodin, P.A, Prince, D.B, Eakin, A.E, Sherer, B.A. | | Deposit date: | 2011-10-03 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Pyrrolamide DNA gyrase inhibitors: fragment-based nuclear magnetic resonance screening to identify antibacterial agents.
Antimicrob.Agents Chemother., 56, 2012
|
|
2BDN
 
 | | Crystal structure of human MCP-1 bound to a blocking antibody, 11K2 | | Descriptor: | Antibody heavy chain 11K2, Antibody light chain 11K2, Small inducible cytokine A2 | | Authors: | Boriack-Sjodin, P.A, Rushe, M, Reid, C, Jarpe, M, van Vlijmen, H, Bailly, V. | | Deposit date: | 2005-10-20 | | Release date: | 2006-06-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structure activity relationships of monocyte chemoattractant proteins in complex with a blocking antibody.
Protein Eng.Des.Sel., 19, 2006
|
|
4EM7
 
 | | Crystal structure of a topoisomerase ATP inhibitor | | Descriptor: | 3-[3-(1H-pyrrolo[2,3-b]pyridin-5-yl)phenyl]propanoic acid, DNA topoisomerase IV, B subunit | | Authors: | Boriack-Sjodin, P.A, Manchester, J. | | Deposit date: | 2012-04-11 | | Release date: | 2012-08-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a novel azaindole class of antibacterial agents targeting the ATPase domains of DNA gyrase and Topoisomerase IV.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4EMV
 
 | | Crystal structure of a topoisomerase ATP inhibitor | | Descriptor: | 5-{2-(ethylcarbamoyl)-4-[3-(trifluoromethyl)-1H-pyrazol-1-yl]-1H-pyrrolo[2,3-b]pyridin-5-yl}pyridine-3-carboxylic acid, DNA topoisomerase IV, B subunit | | Authors: | Boriack-Sjodin, P.A, Manchester, J, Hull, K. | | Deposit date: | 2012-04-12 | | Release date: | 2012-08-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of a novel azaindole class of antibacterial agents targeting the ATPase domains of DNA gyrase and Topoisomerase IV.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4LH7
 
 | | Crystal structure of a LigA inhibitor | | Descriptor: | 4-aminothieno[3,2-c]pyridine-2,7-dicarboxamide, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, ... | | Authors: | Boriack-Sjodin, P.A, Prince, D.B. | | Deposit date: | 2013-06-30 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification through structure-based methods of a bacterial NAD(+)-dependent DNA ligase inhibitor that avoids known resistance mutations.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
5CCM
 
 | | Crystal structure of SMYD3 with SAM and EPZ030456 | | Descriptor: | 6-chloranyl-2-oxidanylidene-N-[(1S,5R)-8-[4-[(phenylmethyl)amino]piperidin-1-yl]sulfonyl-8-azabicyclo[3.2.1]octan-3-yl]-1,3-dihydroindole-5-carboxamide, Histone-lysine N-methyltransferase SMYD3, S-ADENOSYLMETHIONINE, ... | | Authors: | Boriack-Sjodin, P.A. | | Deposit date: | 2015-07-02 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel Oxindole Sulfonamides and Sulfamides: EPZ031686, the First Orally Bioavailable Small Molecule SMYD3 Inhibitor.
Acs Med.Chem.Lett., 7, 2016
|
|
5CCL
 
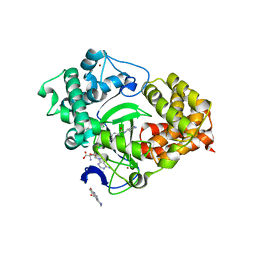 | | Crystal structure of SMYD3 with SAM and oxindole compound | | Descriptor: | 1,2-ETHANEDIOL, 2-oxidanylidene-N-piperidin-4-yl-1,3-dihydroindole-5-carboxamide, Histone-lysine N-methyltransferase SMYD3, ... | | Authors: | Boriack-Sjodin, P.A. | | Deposit date: | 2015-07-02 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Novel Oxindole Sulfonamides and Sulfamides: EPZ031686, the First Orally Bioavailable Small Molecule SMYD3 Inhibitor.
Acs Med.Chem.Lett., 7, 2016
|
|
5EMM
 
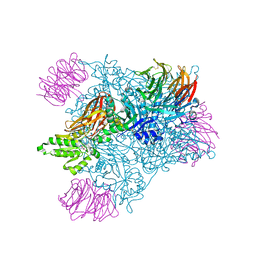 | |
5EML
 
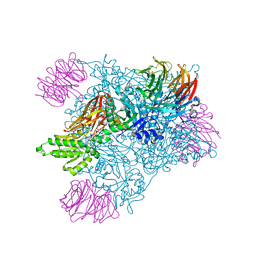 | |
5EMK
 
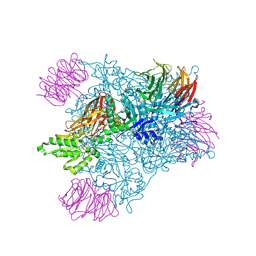 | |
5EMJ
 
 | | Crystal structure of PRMT5:MEP50 with Compound 8 and sinefungin | | Descriptor: | (2~{S})-1-(3,4-dihydro-1~{H}-isoquinolin-2-yl)-3-[[4-(3-methylbenzimidazol-5-yl)pyridin-2-yl]amino]propan-2-ol, GLYCEROL, Methylosome protein 50, ... | | Authors: | Boriack-Sjodin, P.A, Jin, L. | | Deposit date: | 2015-11-06 | | Release date: | 2016-02-24 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (2.273 Å) | | Cite: | Structure and Property Guided Design in the Identification of PRMT5 Tool Compound EPZ015666.
ACS Med Chem Lett, 7, 2016
|
|
1A42
 
 | | HUMAN CARBONIC ANHYDRASE II COMPLEXED WITH BRINZOLAMIDE | | Descriptor: | (4R)-2-(2-ethoxyethyl)-4-(ethylamino)-3,4-dihydro-2H-thieno[3,2-e][1,2]thiazine-6-sulfonamide 1,1-dioxide, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Boriack-Sjodin, P.A, Christianson, D.W. | | Deposit date: | 1998-02-10 | | Release date: | 1999-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of murine carbonic anhydrase IV and human carbonic anhydrase II complexed with brinzolamide: molecular basis of isozyme-drug discrimination.
Protein Sci., 7, 1998
|
|
4W6E
 
 | | Human Tankyrase 1 with small molecule inhibitor | | Descriptor: | 2-(4-{6-[(3S)-3,4-dimethylpiperazin-1-yl]-4-methylpyridin-3-yl}phenyl)-8-(hydroxymethyl)quinazolin-4(3H)-one, Tankyrase-1, ZINC ION | | Authors: | Kazmirski, S.L, Johannes, J, Boriack-Sjodin, P.A, Howard, T. | | Deposit date: | 2014-08-20 | | Release date: | 2015-05-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pyrimidinone nicotinamide mimetics as selective tankyrase and wnt pathway inhibitors suitable for in vivo pharmacology.
Acs Med.Chem.Lett., 6, 2015
|
|
4MOT
 
 | | Structure of Streptococcus pneumonia pare in complex with AZ13072886 | | Descriptor: | 1-[4-(3-methylbutyl)-5-oxo-6-(pyridin-3-yl)-4,5-dihydro[1,3]thiazolo[5,4-b]pyridin-2-yl]-3-prop-2-en-1-ylurea, Topoisomerase IV subunit B | | Authors: | Ogg, D, Boriack-Sjodin, P.A. | | Deposit date: | 2013-09-12 | | Release date: | 2013-11-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Thiazolopyridone ureas as DNA gyrase B inhibitors: Optimization of antitubercular activity and efficacy.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4Y30
 
 | | Crystal structure of human protein arginine methyltransferase PRMT6 bound to SAH and EPZ020411 | | Descriptor: | GLYCEROL, MAGNESIUM ION, N,N'-dimethyl-N-({3-[4-({trans-3-[2-(tetrahydro-2H-pyran-4-yl)ethoxy]cyclobutyl}oxy)phenyl]-1H-pyrazol-4-yl}methyl)ethane-1,2-diamine, ... | | Authors: | Swinger, K.K, Boriack-Sjodin, P.A. | | Deposit date: | 2015-02-10 | | Release date: | 2015-04-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Aryl Pyrazoles as Potent Inhibitors of Arginine Methyltransferases: Identification of the First PRMT6 Tool Compound.
Acs Med.Chem.Lett., 6, 2015
|
|
4XDO
 
 | | Crystal structure of human KDM4C catalytic domain with OGA | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Lysine-specific demethylase 4C, ... | | Authors: | Swinger, K.K, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-19 | | Release date: | 2015-03-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A High-Throughput Mass Spectrometry Assay Coupled with Redox Activity Testing Reduces Artifacts and False Positives in Lysine Demethylase Screening.
J Biomol Screen, 20, 2015
|
|
4Y2H
 
 | |
8SZS
 
 | | Cat DHX9 bound to GDP | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lee, Y.-T, Sickmier, E.A, Boriack-Sjodin, P.A. | | Deposit date: | 2023-05-30 | | Release date: | 2023-09-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Development of assays to support identification and characterization of modulators of DExH-box helicase DHX9.
Slas Discov, 28, 2023
|
|
